Anne-Christin Hauschild
S-Chain: Structured Visual Chain-of-Thought For Medicine
Oct 26, 2025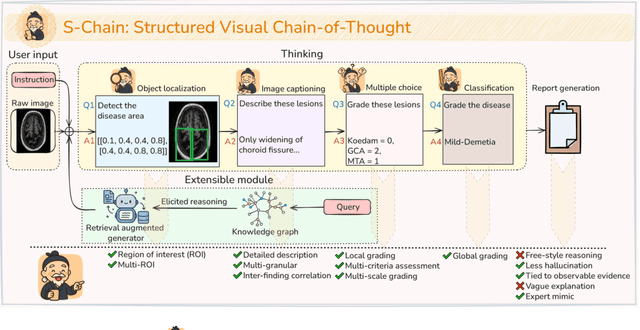
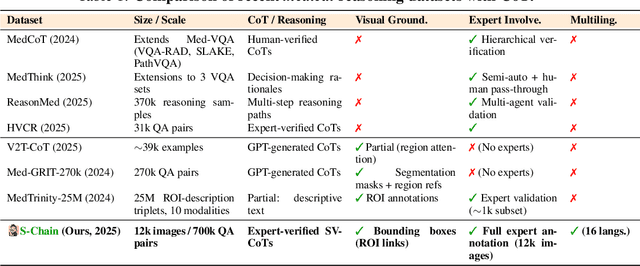
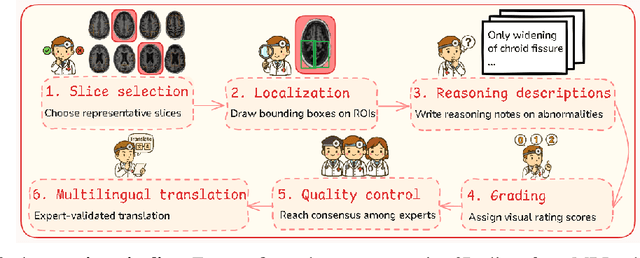
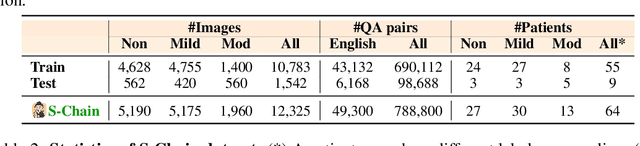
Abstract:Faithful reasoning in medical vision-language models (VLMs) requires not only accurate predictions but also transparent alignment between textual rationales and visual evidence. While Chain-of-Thought (CoT) prompting has shown promise in medical visual question answering (VQA), no large-scale expert-level dataset has captured stepwise reasoning with precise visual grounding. We introduce S-Chain, the first large-scale dataset of 12,000 expert-annotated medical images with bounding boxes and structured visual CoT (SV-CoT), explicitly linking visual regions to reasoning steps. The dataset further supports 16 languages, totaling over 700k VQA pairs for broad multilingual applicability. Using S-Chain, we benchmark state-of-the-art medical VLMs (ExGra-Med, LLaVA-Med) and general-purpose VLMs (Qwen2.5-VL, InternVL2.5), showing that SV-CoT supervision significantly improves interpretability, grounding fidelity, and robustness. Beyond benchmarking, we study its synergy with retrieval-augmented generation, revealing how domain knowledge and visual grounding interact during autoregressive reasoning. Finally, we propose a new mechanism that strengthens the alignment between visual evidence and reasoning, improving both reliability and efficiency. S-Chain establishes a new benchmark for grounded medical reasoning and paves the way toward more trustworthy and explainable medical VLMs.
MGPATH: Vision-Language Model with Multi-Granular Prompt Learning for Few-Shot WSI Classification
Feb 11, 2025



Abstract:Whole slide pathology image classification presents challenges due to gigapixel image sizes and limited annotation labels, hindering model generalization. This paper introduces a prompt learning method to adapt large vision-language models for few-shot pathology classification. We first extend the Prov-GigaPath vision foundation model, pre-trained on 1.3 billion pathology image tiles, into a vision-language model by adding adaptors and aligning it with medical text encoders via contrastive learning on 923K image-text pairs. The model is then used to extract visual features and text embeddings from few-shot annotations and fine-tunes with learnable prompt embeddings. Unlike prior methods that combine prompts with frozen features using prefix embeddings or self-attention, we propose multi-granular attention that compares interactions between learnable prompts with individual image patches and groups of them. This approach improves the model's ability to capture both fine-grained details and broader context, enhancing its recognition of complex patterns across sub-regions. To further improve accuracy, we leverage (unbalanced) optimal transport-based visual-text distance to secure model robustness by mitigating perturbations that might occur during the data augmentation process. Empirical experiments on lung, kidney, and breast pathology modalities validate the effectiveness of our approach; thereby, we surpass several of the latest competitors and consistently improve performance across diverse architectures, including CLIP, PLIP, and Prov-GigaPath integrated PLIP. We release our implementations and pre-trained models at this MGPATH.
Federated Random Forest for Partially Overlapping Clinical Data
May 31, 2024



Abstract:In the healthcare sector, a consciousness surrounding data privacy and corresponding data protection regulations, as well as heterogeneous and non-harmonized data, pose huge challenges to large-scale data analysis. Moreover, clinical data often involves partially overlapping features, as some observations may be missing due to various reasons, such as differences in procedures, diagnostic tests, or other recorded patient history information across hospitals or institutes. To address the challenges posed by partially overlapping features and incomplete data in clinical datasets, a comprehensive approach is required. Particularly in the domain of medical data, promising outcomes are achieved by federated random forests whenever features align. However, for most standard algorithms, like random forest, it is essential that all data sets have identical parameters. Therefore, in this work the concept of federated random forest is adapted to a setting with partially overlapping features. Moreover, our research assesses the effectiveness of the newly developed federated random forest models for partially overlapping clinical data. For aggregating the federated, globally optimized model, only features available locally at each site can be used. We tackled two issues in federation: (i) the quantity of involved parties, (ii) the varying overlap of features. This evaluation was conducted across three clinical datasets. The federated random forest model even in cases where only a subset of features overlaps consistently demonstrates superior performance compared to its local counterpart. This holds true across various scenarios, including datasets with imbalanced classes. Consequently, federated random forests for partially overlapped data offer a promising solution to transcend barriers in collaborative research and corporate cooperation.
Analysis of a Deep Learning Model for 12-Lead ECG Classification Reveals Learned Features Similar to Diagnostic Criteria
Nov 03, 2022



Abstract:Despite their remarkable performance, deep neural networks remain unadopted in clinical practice, which is considered to be partially due to their lack in explainability. In this work, we apply attribution methods to a pre-trained deep neural network (DNN) for 12-lead electrocardiography classification to open this "black box" and understand the relationship between model prediction and learned features. We classify data from a public data set and the attribution methods assign a "relevance score" to each sample of the classified signals. This allows analyzing what the network learned during training, for which we propose quantitative methods: average relevance scores over a) classes, b) leads, and c) average beats. The analyses of relevance scores for atrial fibrillation (AF) and left bundle branch block (LBBB) compared to healthy controls show that their mean values a) increase with higher classification probability and correspond to false classifications when around zero, and b) correspond to clinical recommendations regarding which lead to consider. Furthermore, c) visible P-waves and concordant T-waves result in clearly negative relevance scores in AF and LBBB classification, respectively. In summary, our analysis suggests that the DNN learned features similar to cardiology textbook knowledge.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
May 12, 2021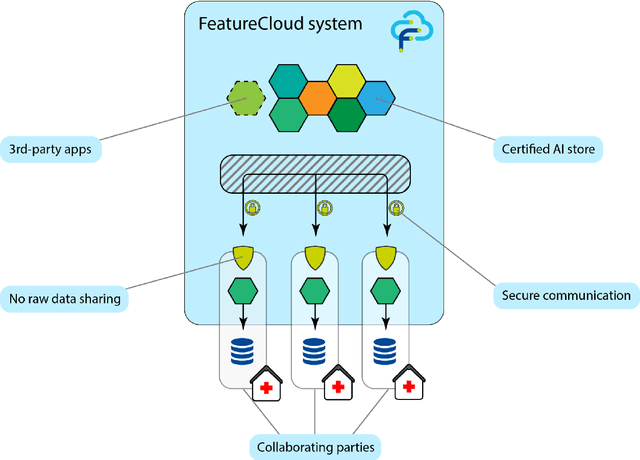
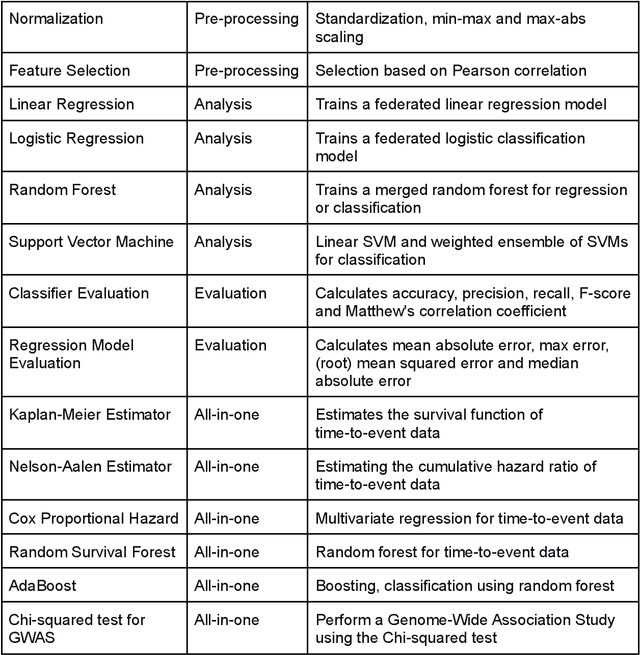
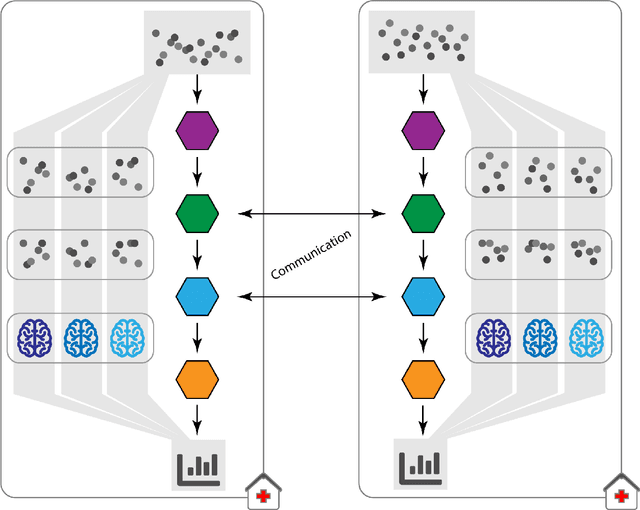
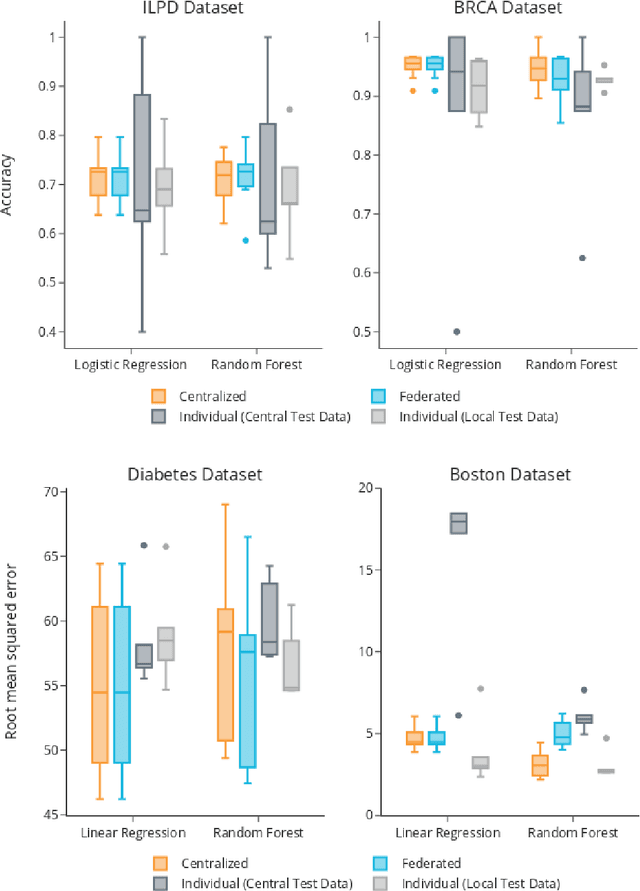
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge