Duy Minh Ho Nguyen
DuFal: Dual-Frequency-Aware Learning for High-Fidelity Extremely Sparse-view CBCT Reconstruction
Jan 21, 2026Abstract:Sparse-view Cone-Beam Computed Tomography reconstruction from limited X-ray projections remains a challenging problem in medical imaging due to the inherent undersampling of fine-grained anatomical details, which correspond to high-frequency components. Conventional CNN-based methods often struggle to recover these fine structures, as they are typically biased toward learning low-frequency information. To address this challenge, this paper presents DuFal (Dual-Frequency-Aware Learning), a novel framework that integrates frequency-domain and spatial-domain processing via a dual-path architecture. The core innovation lies in our High-Local Factorized Fourier Neural Operator, which comprises two complementary branches: a Global High-Frequency Enhanced Fourier Neural Operator that captures global frequency patterns and a Local High-Frequency Enhanced Fourier Neural Operator that processes spatially partitioned patches to preserve spatial locality that might be lost in global frequency analysis. To improve efficiency, we design a Spectral-Channel Factorization scheme that reduces the Fourier Neural Operator parameter count. We also design a Cross-Attention Frequency Fusion module to integrate spatial and frequency features effectively. The fused features are then decoded through a Feature Decoder to produce projection representations, which are subsequently processed through an Intensity Field Decoding pipeline to reconstruct a final Computed Tomography volume. Experimental results on the LUNA16 and ToothFairy datasets demonstrate that DuFal significantly outperforms existing state-of-the-art methods in preserving high-frequency anatomical features, particularly under extremely sparse-view settings.
Rethinking Progression of Memory State in Robotic Manipulation: An Object-Centric Perspective
Nov 18, 2025Abstract:As embodied agents operate in increasingly complex environments, the ability to perceive, track, and reason about individual object instances over time becomes essential, especially in tasks requiring sequenced interactions with visually similar objects. In these non-Markovian settings, key decision cues are often hidden in object-specific histories rather than the current scene. Without persistent memory of prior interactions (what has been interacted with, where it has been, or how it has changed) visuomotor policies may fail, repeat past actions, or overlook completed ones. To surface this challenge, we introduce LIBERO-Mem, a non-Markovian task suite for stress-testing robotic manipulation under object-level partial observability. It combines short- and long-horizon object tracking with temporally sequenced subgoals, requiring reasoning beyond the current frame. However, vision-language-action (VLA) models often struggle in such settings, with token scaling quickly becoming intractable even for tasks spanning just a few hundred frames. We propose Embodied-SlotSSM, a slot-centric VLA framework built for temporal scalability. It maintains spatio-temporally consistent slot identities and leverages them through two mechanisms: (1) slot-state-space modeling for reconstructing short-term history, and (2) a relational encoder to align the input tokens with action decoding. Together, these components enable temporally grounded, context-aware action prediction. Experiments show Embodied-SlotSSM's baseline performance on LIBERO-Mem and general tasks, offering a scalable solution for non-Markovian reasoning in object-centric robotic policies.
MGPATH: Vision-Language Model with Multi-Granular Prompt Learning for Few-Shot WSI Classification
Feb 11, 2025



Abstract:Whole slide pathology image classification presents challenges due to gigapixel image sizes and limited annotation labels, hindering model generalization. This paper introduces a prompt learning method to adapt large vision-language models for few-shot pathology classification. We first extend the Prov-GigaPath vision foundation model, pre-trained on 1.3 billion pathology image tiles, into a vision-language model by adding adaptors and aligning it with medical text encoders via contrastive learning on 923K image-text pairs. The model is then used to extract visual features and text embeddings from few-shot annotations and fine-tunes with learnable prompt embeddings. Unlike prior methods that combine prompts with frozen features using prefix embeddings or self-attention, we propose multi-granular attention that compares interactions between learnable prompts with individual image patches and groups of them. This approach improves the model's ability to capture both fine-grained details and broader context, enhancing its recognition of complex patterns across sub-regions. To further improve accuracy, we leverage (unbalanced) optimal transport-based visual-text distance to secure model robustness by mitigating perturbations that might occur during the data augmentation process. Empirical experiments on lung, kidney, and breast pathology modalities validate the effectiveness of our approach; thereby, we surpass several of the latest competitors and consistently improve performance across diverse architectures, including CLIP, PLIP, and Prov-GigaPath integrated PLIP. We release our implementations and pre-trained models at this MGPATH.
On the Out of Distribution Robustness of Foundation Models in Medical Image Segmentation
Nov 18, 2023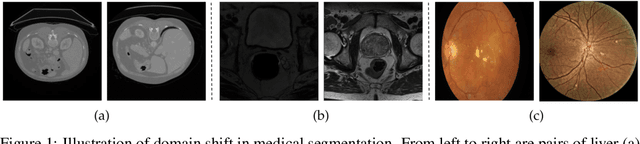
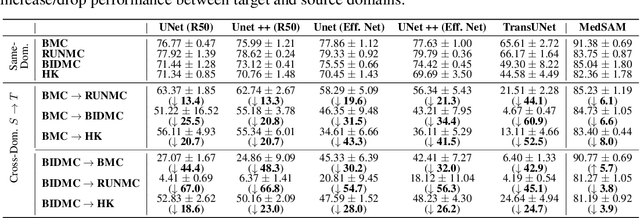
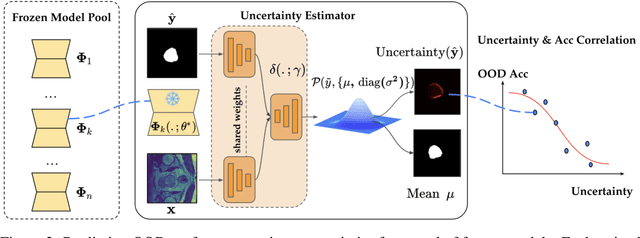
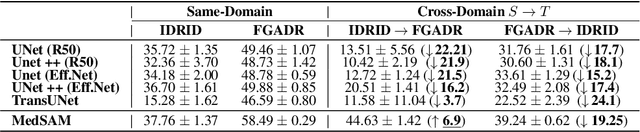
Abstract:Constructing a robust model that can effectively generalize to test samples under distribution shifts remains a significant challenge in the field of medical imaging. The foundational models for vision and language, pre-trained on extensive sets of natural image and text data, have emerged as a promising approach. It showcases impressive learning abilities across different tasks with the need for only a limited amount of annotated samples. While numerous techniques have focused on developing better fine-tuning strategies to adapt these models for specific domains, we instead examine their robustness to domain shifts in the medical image segmentation task. To this end, we compare the generalization performance to unseen domains of various pre-trained models after being fine-tuned on the same in-distribution dataset and show that foundation-based models enjoy better robustness than other architectures. From here, we further developed a new Bayesian uncertainty estimation for frozen models and used them as an indicator to characterize the model's performance on out-of-distribution (OOD) data, proving particularly beneficial for real-world applications. Our experiments not only reveal the limitations of current indicators like accuracy on the line or agreement on the line commonly used in natural image applications but also emphasize the promise of the introduced Bayesian uncertainty. Specifically, lower uncertainty predictions usually tend to higher out-of-distribution (OOD) performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge