Anne Hartebrodt
UnPaSt: unsupervised patient stratification by differentially expressed biclusters in omics data
Jul 31, 2024Abstract:Most complex diseases, including cancer and non-malignant diseases like asthma, have distinct molecular subtypes that require distinct clinical approaches. However, existing computational patient stratification methods have been benchmarked almost exclusively on cancer omics data and only perform well when mutually exclusive subtypes can be characterized by many biomarkers. Here, we contribute with a massive evaluation attempt, quantitatively exploring the power of 22 unsupervised patient stratification methods using both, simulated and real transcriptome data. From this experience, we developed UnPaSt (https://apps.cosy.bio/unpast/) optimizing unsupervised patient stratification, working even with only a limited number of subtype-predictive biomarkers. We evaluated all 23 methods on real-world breast cancer and asthma transcriptomics data. Although many methods reliably detected major breast cancer subtypes, only few identified Th2-high asthma, and UnPaSt significantly outperformed its closest competitors in both test datasets. Essentially, we showed that UnPaSt can detect many biologically insightful and reproducible patterns in omic datasets.
Privacy-Preserving Multi-Center Differential Protein Abundance Analysis with FedProt
Jul 21, 2024



Abstract:Quantitative mass spectrometry has revolutionized proteomics by enabling simultaneous quantification of thousands of proteins. Pooling patient-derived data from multiple institutions enhances statistical power but raises significant privacy concerns. Here we introduce FedProt, the first privacy-preserving tool for collaborative differential protein abundance analysis of distributed data, which utilizes federated learning and additive secret sharing. In the absence of a multicenter patient-derived dataset for evaluation, we created two, one at five centers from LFQ E.coli experiments and one at three centers from TMT human serum. Evaluations using these datasets confirm that FedProt achieves accuracy equivalent to DEqMS applied to pooled data, with completely negligible absolute differences no greater than $\text{$4 \times 10^{-12}$}$. In contrast, -log10(p-values) computed by the most accurate meta-analysis methods diverged from the centralized analysis results by up to 25-27. FedProt is available as a web tool with detailed documentation as a FeatureCloud App.
Privacy of federated QR decomposition using additive secure multiparty computation
Oct 12, 2022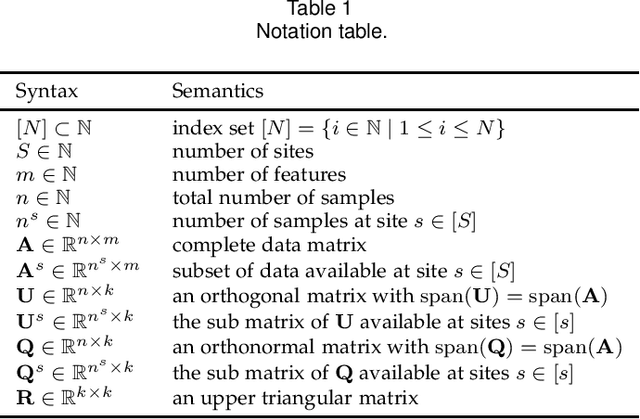
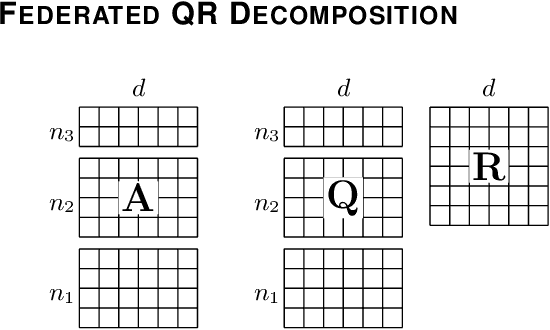
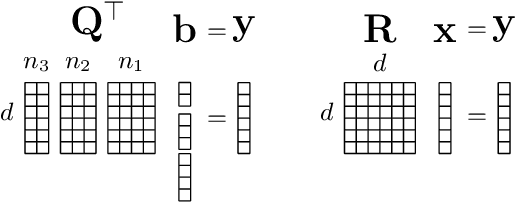
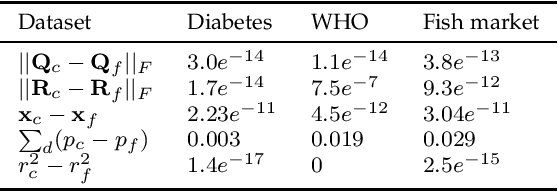
Abstract:Federated learning (FL) is a privacy-aware data mining strategy keeping the private data on the owners' machine and thereby confidential. The clients compute local models and send them to an aggregator which computes a global model. In hybrid FL, the local parameters are additionally masked using secure aggregation, such that only the global aggregated statistics become available in clear text, not the client specific updates. Federated QR decomposition has not been studied extensively in the context of cross-silo federated learning. In this article, we investigate the suitability of three QR decomposition algorithms for cross-silo FL and suggest a privacy-aware QR decomposition scheme based on the Gram-Schmidt algorithm which does not blatantly leak raw data. We apply the algorithm to compute linear regression in a federated manner.
Federated singular value decomposition for high dimensional data
May 24, 2022



Abstract:Federated learning (FL) is emerging as a privacy-aware alternative to classical cloud-based machine learning. In FL, the sensitive data remains in data silos and only aggregated parameters are exchanged. Hospitals and research institutions which are not willing to share their data can join a federated study without breaching confidentiality. In addition to the extreme sensitivity of biomedical data, the high dimensionality poses a challenge in the context of federated genome-wide association studies (GWAS). In this article, we present a federated singular value decomposition (SVD) algorithm, suitable for the privacy-related and computational requirements of GWAS. Notably, the algorithm has a transmission cost independent of the number of samples and is only weakly dependent on the number of features, because the singular vectors associated with the samples are never exchanged and the vectors associated with the features only for a fixed number of iterations. Although motivated by GWAS, the algorithm is generically applicable for both horizontally and vertically partitioned data.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
May 12, 2021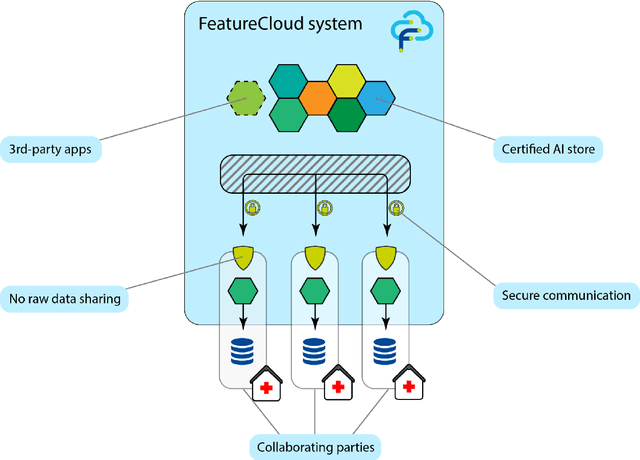
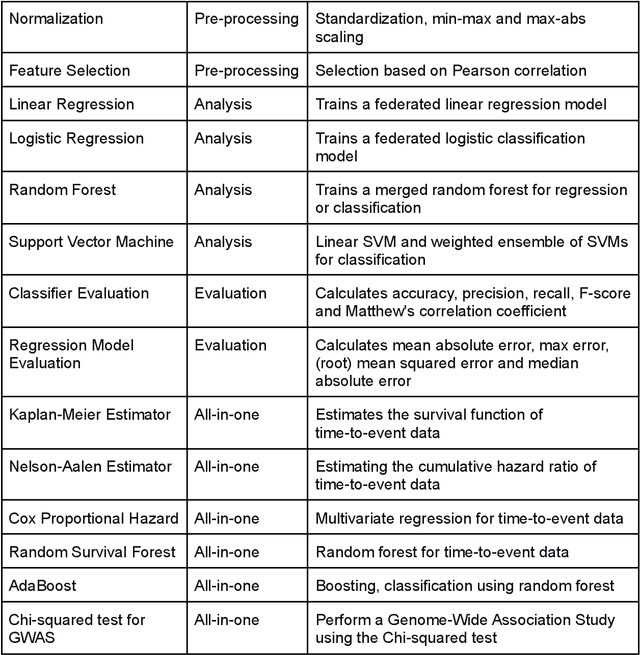
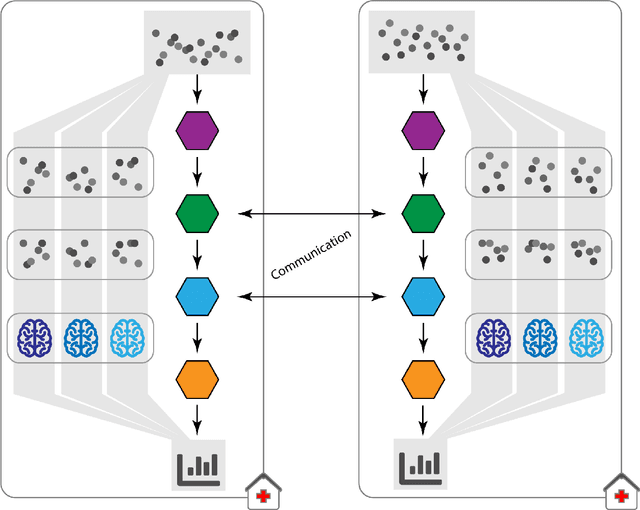
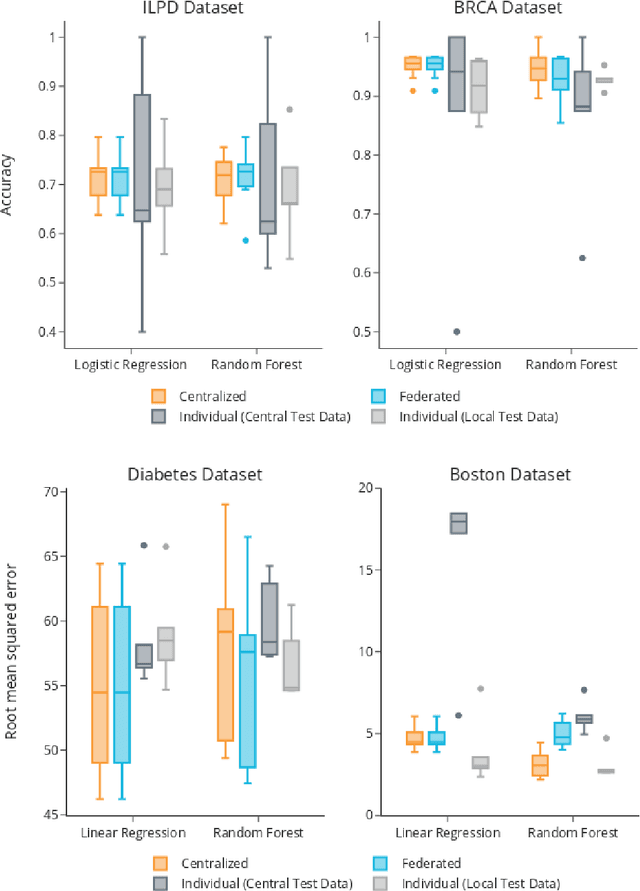
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
Privacy-preserving Artificial Intelligence Techniques in Biomedicine
Jul 22, 2020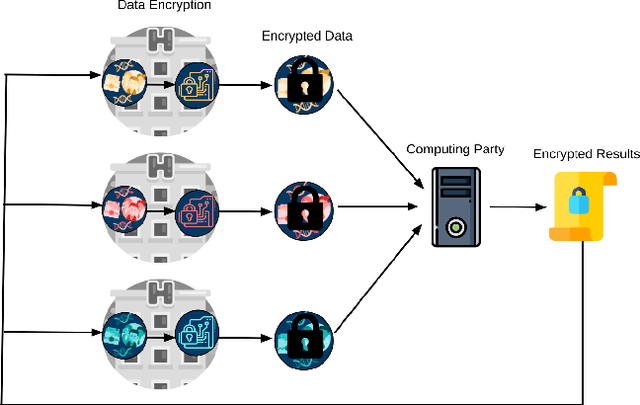
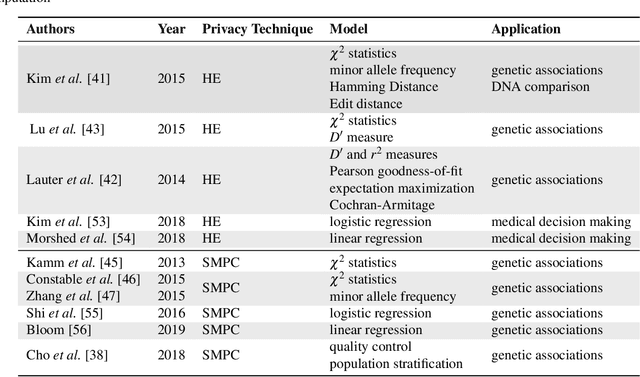
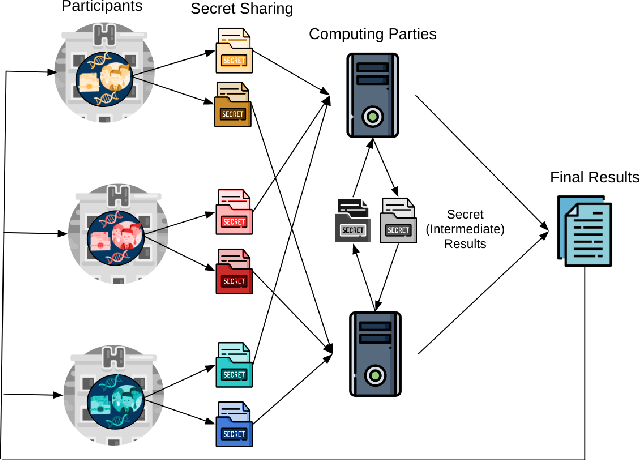
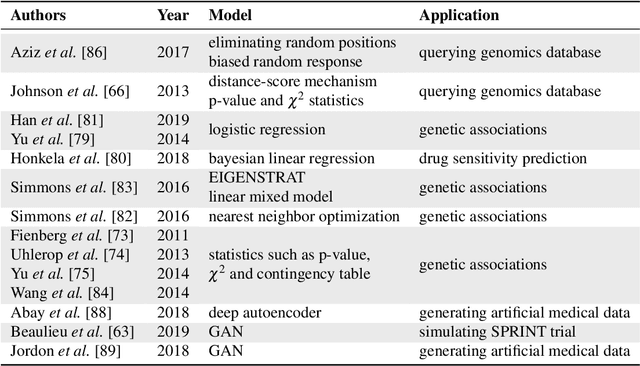
Abstract:Artificial intelligence (AI) has been successfully applied in numerous scientific domains including biomedicine and healthcare. Here, it has led to several breakthroughs ranging from clinical decision support systems, image analysis to whole genome sequencing. However, training an AI model on sensitive data raises also concerns about the privacy of individual participants. Adversary AIs, for example, can abuse even summary statistics of a study to determine the presence or absence of an individual in a given dataset. This has resulted in increasing restrictions to access biomedical data, which in turn is detrimental for collaborative research and impedes scientific progress. Hence there has been an explosive growth in efforts to harness the power of AI for learning from sensitive data while protecting patients' privacy. This paper provides a structured overview of recent advances in privacy-preserving AI techniques in biomedicine. It places the most important state-of-the-art approaches within a unified taxonomy, and discusses their strengths, limitations, and open problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge