David B. Blumenthal
UnPaSt: unsupervised patient stratification by differentially expressed biclusters in omics data
Jul 31, 2024Abstract:Most complex diseases, including cancer and non-malignant diseases like asthma, have distinct molecular subtypes that require distinct clinical approaches. However, existing computational patient stratification methods have been benchmarked almost exclusively on cancer omics data and only perform well when mutually exclusive subtypes can be characterized by many biomarkers. Here, we contribute with a massive evaluation attempt, quantitatively exploring the power of 22 unsupervised patient stratification methods using both, simulated and real transcriptome data. From this experience, we developed UnPaSt (https://apps.cosy.bio/unpast/) optimizing unsupervised patient stratification, working even with only a limited number of subtype-predictive biomarkers. We evaluated all 23 methods on real-world breast cancer and asthma transcriptomics data. Although many methods reliably detected major breast cancer subtypes, only few identified Th2-high asthma, and UnPaSt significantly outperformed its closest competitors in both test datasets. Essentially, we showed that UnPaSt can detect many biologically insightful and reproducible patterns in omic datasets.
Federated singular value decomposition for high dimensional data
May 24, 2022



Abstract:Federated learning (FL) is emerging as a privacy-aware alternative to classical cloud-based machine learning. In FL, the sensitive data remains in data silos and only aggregated parameters are exchanged. Hospitals and research institutions which are not willing to share their data can join a federated study without breaching confidentiality. In addition to the extreme sensitivity of biomedical data, the high dimensionality poses a challenge in the context of federated genome-wide association studies (GWAS). In this article, we present a federated singular value decomposition (SVD) algorithm, suitable for the privacy-related and computational requirements of GWAS. Notably, the algorithm has a transmission cost independent of the number of samples and is only weakly dependent on the number of features, because the singular vectors associated with the samples are never exchanged and the vectors associated with the features only for a fixed number of iterations. Although motivated by GWAS, the algorithm is generically applicable for both horizontally and vertically partitioned data.
The Minimum Edit Arborescence Problem and Its Use in Compressing Graph Collections
Jul 30, 2021
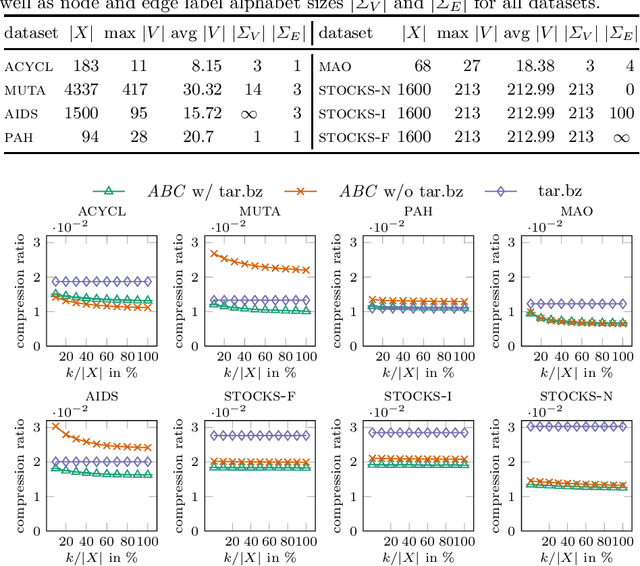

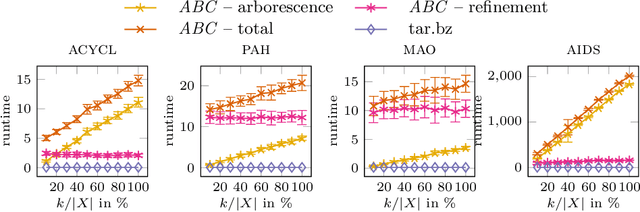
Abstract:The inference of minimum spanning arborescences within a set of objects is a general problem which translates into numerous application-specific unsupervised learning tasks. We introduce a unified and generic structure called edit arborescence that relies on edit paths between data in a collection, as well as the Min Edit Arborescence Problem, which asks for an edit arborescence that minimizes the sum of costs of its inner edit paths. Through the use of suitable cost functions, this generic framework allows to model a variety of problems. In particular, we show that by introducing encoding size preserving edit costs, it can be used as an efficient method for compressing collections of labeled graphs. Experiments on various graph datasets, with comparisons to standard compression tools, show the potential of our method.
Federated Multi-Mini-Batch: An Efficient Training Approach to Federated Learning in Non-IID Environments
Nov 13, 2020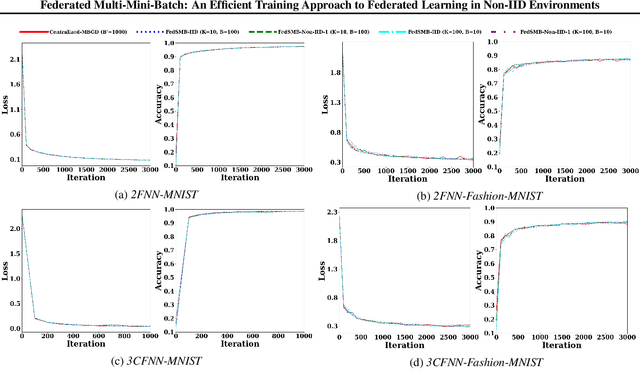
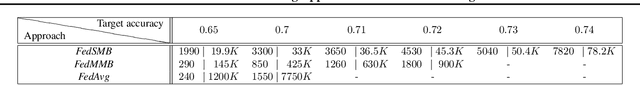
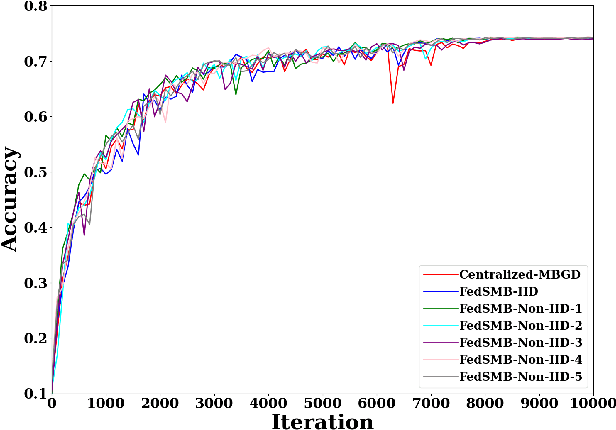
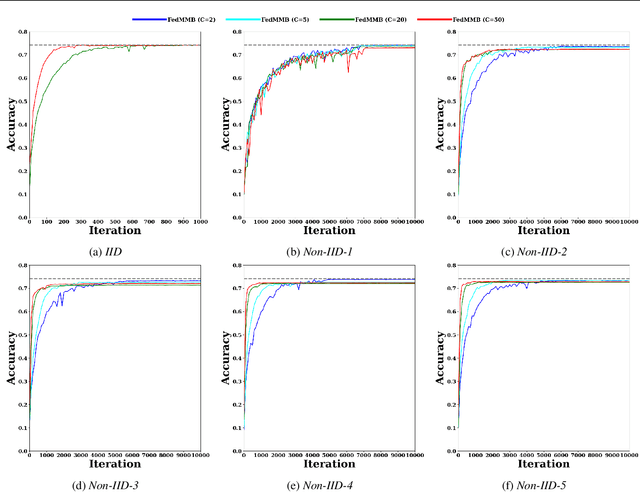
Abstract:Federated learning is a well-established approach to privacy-preserving training of a joint model on heavily distributed data. Federated averaging (FedAvg) is a well-known communication-efficient algorithm for federated learning, which performs well if the data distribution across the clients is independently and identically distributed (IID). However, FedAvg provides a lower accuracy and still requires a large number of communication rounds to achieve a target accuracy when it comes to Non-IID environments. To address the former limitation, we present federated single mini-batch (FedSMB), where the clients train the model on a single mini-batch from their dataset in each iteration. We show that FedSMB achieves the accuracy of the centralized training in Non-IID configurations, but in a considerable number of iterations. To address the latter limitation, we introduce federated multi-mini-batch (FedMMB) as a generalization of FedSMB, where the clients train the model on multiple mini-batches (specified by the batch count) in each communication round. FedMMB decouples the batch size from the batch count and provides a trade-off between the accuracy and communication efficiency in Non-IID settings. This is not possible with FedAvg, in which a single parameter determines both the batch size and batch count. The simulation results illustrate that FedMMB outperforms FedAvg in terms of the accuracy, communication efficiency, as well as computational efficiency and is an efficient training approach to federated learning in Non-IID environments.
Privacy-preserving Artificial Intelligence Techniques in Biomedicine
Jul 22, 2020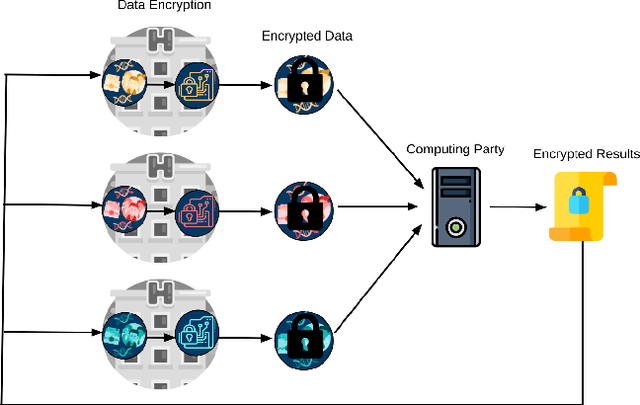
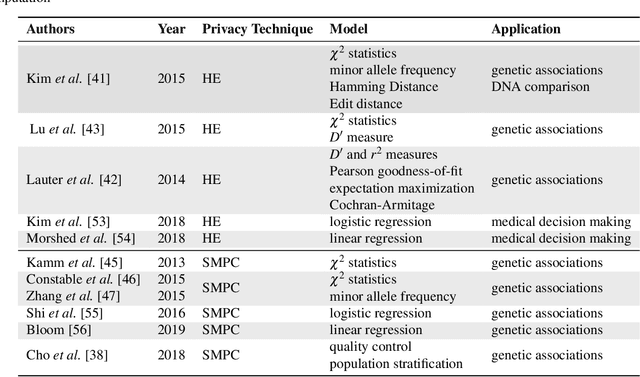
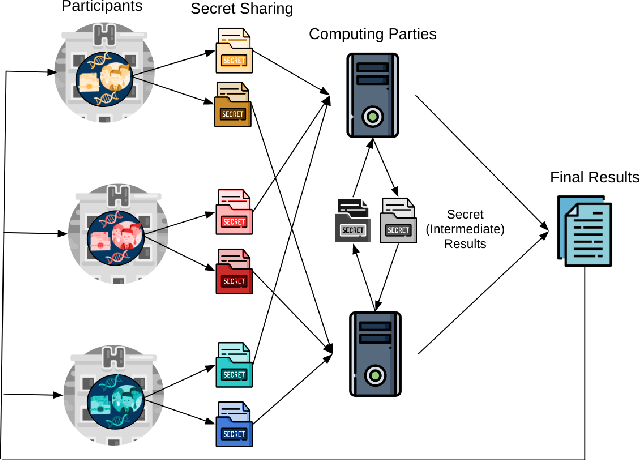
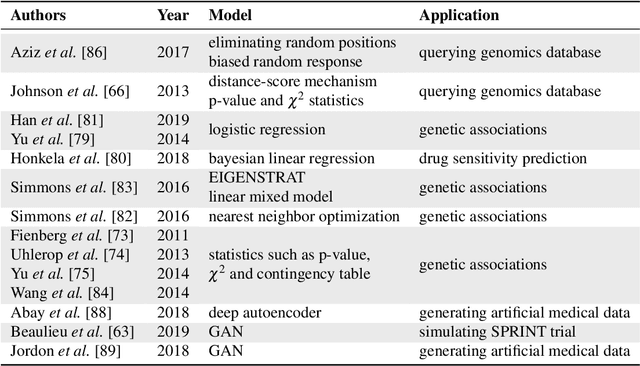
Abstract:Artificial intelligence (AI) has been successfully applied in numerous scientific domains including biomedicine and healthcare. Here, it has led to several breakthroughs ranging from clinical decision support systems, image analysis to whole genome sequencing. However, training an AI model on sensitive data raises also concerns about the privacy of individual participants. Adversary AIs, for example, can abuse even summary statistics of a study to determine the presence or absence of an individual in a given dataset. This has resulted in increasing restrictions to access biomedical data, which in turn is detrimental for collaborative research and impedes scientific progress. Hence there has been an explosive growth in efforts to harness the power of AI for learning from sensitive data while protecting patients' privacy. This paper provides a structured overview of recent advances in privacy-preserving AI techniques in biomedicine. It places the most important state-of-the-art approaches within a unified taxonomy, and discusses their strengths, limitations, and open problems.
New Techniques for Graph Edit Distance Computation
Aug 01, 2019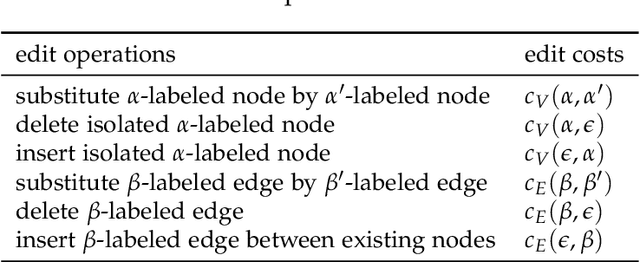
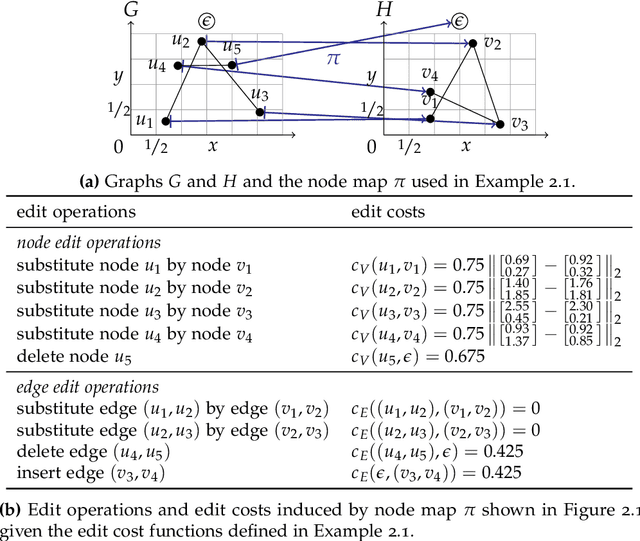
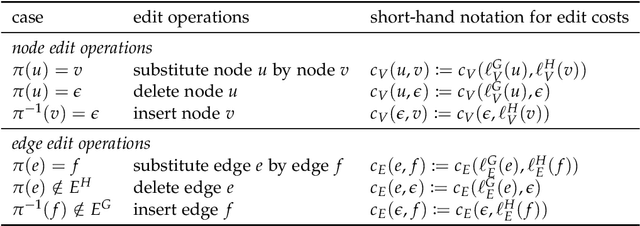
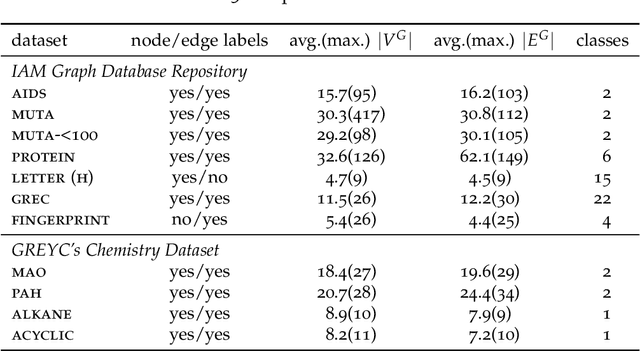
Abstract:Due to their capacity to encode rich structural information, labeled graphs are often used for modeling various kinds of objects such as images, molecules, and chemical compounds. If pattern recognition problems such as clustering and classification are to be solved on these domains, a (dis-)similarity measure for labeled graphs has to be defined. A widely used measure is the graph edit distance (GED), which, intuitively, is defined as the minimum amount of distortion that has to be applied to a source graph in order to transform it into a target graph. The main advantage of GED is its flexibility and sensitivity to small differences between the input graphs. Its main drawback is that it is hard to compute. In this thesis, new results and techniques for several aspects of computing GED are presented. Firstly, theoretical aspects are discussed: competing definitions of GED are harmonized, the problem of computing GED is characterized in terms of complexity, and several reductions from GED to the quadratic assignment problem (QAP) are presented. Secondly, solvers for the linear sum assignment problem with error-correction (LSAPE) are discussed. LSAPE is a generalization of the well-known linear sum assignment problem (LSAP), and has to be solved as a subproblem by many GED algorithms. In particular, a new solver is presented that efficiently reduces LSAPE to LSAP. Thirdly, exact algorithms for computing GED are presented in a systematic way, and improvements of existing algorithms as well as a new mixed integer programming (MIP) based approach are introduced. Fourthly, a detailed overview of heuristic algorithms that approximate GED via upper and lower bounds is provided, and eight new heuristics are described. Finally, a new easily extensible C++ library for exactly or approximately computing GED is presented.
Improved local search for graph edit distance
Jul 05, 2019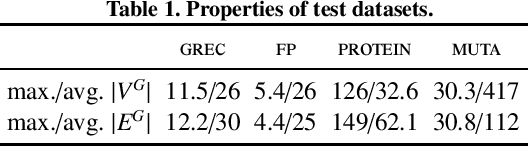



Abstract:Graph Edit Distance (GED) measures the dissimilarity between two graphs as the minimal cost of a sequence of elementary operations transforming one graph into another. This measure is fundamental in many areas such as structural pattern recognition or classification. However, exactly computing GED is NP-hard. Among different classes of heuristic algorithms that were proposed to compute approximate solutions, local search based algorithms provide the tightest upper bounds for GED. In this paper, we present K-REFINE and RANDPOST. K-REFINE generalizes and improves an existing local search algorithm and performs particularly well on small graphs. RANDPOST is a general warm start framework that stochastically generates promising initial solutions to be used by any local search based GED algorithm. It is particularly efficient on large graphs. An extensive empirical evaluation demonstrates that both K-REFINE and RANDPOST perform excellently in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge