Jan Baumbach
FTA-FTL: A Fine-Tuned Aggregation Federated Transfer Learning Scheme for Lithology Microscopic Image Classification
Jan 06, 2025



Abstract:Lithology discrimination is a crucial activity in characterizing oil reservoirs, and processing lithology microscopic images is an essential technique for investigating fossils and minerals and geological assessment of shale oil exploration. In this way, Deep Learning (DL) technique is a powerful approach for building robust classifier models. However, there is still a considerable challenge to collect and produce a large dataset. Transfer-learning and data augmentation techniques have emerged as popular approaches to tackle this problem. Furthermore, due to different reasons, especially data privacy, individuals, organizations, and industry companies often are not willing to share their sensitive data and information. Federated Learning (FL) has emerged to train a highly accurate central model across multiple decentralized edge servers without transferring sensitive data, preserving sensitive data, and enhancing security. This study involves two phases; the first phase is to conduct Lithology microscopic image classification on a small dataset using transfer learning. In doing so, various pre-trained DL model architectures are comprehensively compared for the classification task. In the second phase, we formulated the classification task to a Federated Transfer Learning (FTL) scheme and proposed a Fine-Tuned Aggregation strategy for Federated Learning (FTA-FTL). In order to perform a comprehensive experimental study, several metrics such as accuracy, f1 score, precision, specificity, sensitivity (recall), and confusion matrix are taken into account. The results are in excellent agreement and confirm the efficiency of the proposed scheme, and show that the proposed FTA-FTL algorithm is capable enough to achieve approximately the same results obtained by the centralized implementation for Lithology microscopic images classification task.
FedRBE -- a decentralized privacy-preserving federated batch effect correction tool for omics data based on limma
Dec 08, 2024Abstract:Batch effects in omics data obscure true biological signals and constitute a major challenge for privacy-preserving analyses of distributed patient data. Existing batch effect correction methods either require data centralization, which may easily conflict with privacy requirements, or lack support for missing values and automated workflows. To bridge this gap, we developed fedRBE, a federated implementation of limma's removeBatchEffect method. We implemented it as an app for the FeatureCloud platform. Unlike its existing analogs, fedRBE effectively handles data with missing values and offers an automated, user-friendly online user interface (https://featurecloud.ai/app/fedrbe). Leveraging secure multi-party computation provides enhanced security guarantees over classical federated learning approaches. We evaluated our fedRBE algorithm on simulated and real omics data, achieving performance comparable to the centralized method with negligible differences (no greater than 3.6E-13). By enabling collaborative correction without data sharing, fedRBE facilitates large-scale omics studies where batch effect correction is crucial.
UnPaSt: unsupervised patient stratification by differentially expressed biclusters in omics data
Jul 31, 2024Abstract:Most complex diseases, including cancer and non-malignant diseases like asthma, have distinct molecular subtypes that require distinct clinical approaches. However, existing computational patient stratification methods have been benchmarked almost exclusively on cancer omics data and only perform well when mutually exclusive subtypes can be characterized by many biomarkers. Here, we contribute with a massive evaluation attempt, quantitatively exploring the power of 22 unsupervised patient stratification methods using both, simulated and real transcriptome data. From this experience, we developed UnPaSt (https://apps.cosy.bio/unpast/) optimizing unsupervised patient stratification, working even with only a limited number of subtype-predictive biomarkers. We evaluated all 23 methods on real-world breast cancer and asthma transcriptomics data. Although many methods reliably detected major breast cancer subtypes, only few identified Th2-high asthma, and UnPaSt significantly outperformed its closest competitors in both test datasets. Essentially, we showed that UnPaSt can detect many biologically insightful and reproducible patterns in omic datasets.
Privacy-Preserving Multi-Center Differential Protein Abundance Analysis with FedProt
Jul 21, 2024



Abstract:Quantitative mass spectrometry has revolutionized proteomics by enabling simultaneous quantification of thousands of proteins. Pooling patient-derived data from multiple institutions enhances statistical power but raises significant privacy concerns. Here we introduce FedProt, the first privacy-preserving tool for collaborative differential protein abundance analysis of distributed data, which utilizes federated learning and additive secret sharing. In the absence of a multicenter patient-derived dataset for evaluation, we created two, one at five centers from LFQ E.coli experiments and one at three centers from TMT human serum. Evaluations using these datasets confirm that FedProt achieves accuracy equivalent to DEqMS applied to pooled data, with completely negligible absolute differences no greater than $\text{$4 \times 10^{-12}$}$. In contrast, -log10(p-values) computed by the most accurate meta-analysis methods diverged from the centralized analysis results by up to 25-27. FedProt is available as a web tool with detailed documentation as a FeatureCloud App.
HyFed: A Hybrid Federated Framework for Privacy-preserving Machine Learning
May 21, 2021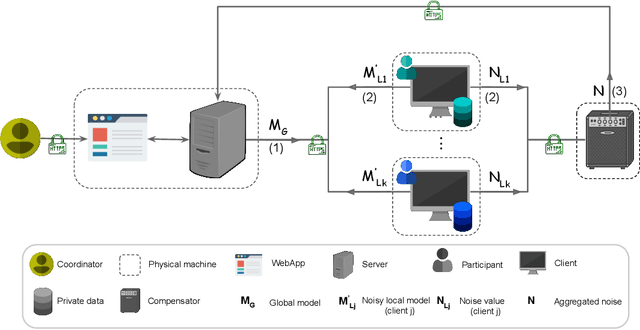

Abstract:Federated learning (FL) enables multiple clients to jointly train a global model under the coordination of a central server. Although FL is a privacy-aware paradigm, where raw data sharing is not required, recent studies have shown that FL might leak the private data of a client through the model parameters shared with the server or the other clients. In this paper, we present the HyFed framework, which enhances the privacy of FL while preserving the utility of the global model. HyFed provides developers with a generic API to develop federated, privacy-preserving algorithms. HyFed supports both simulation and federated operation modes and its source code is publicly available at https://github.com/tum-aimed/hyfed.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
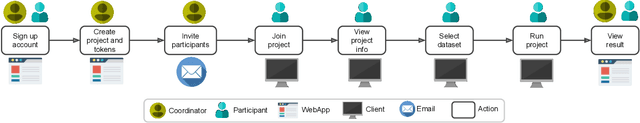
May 12, 2021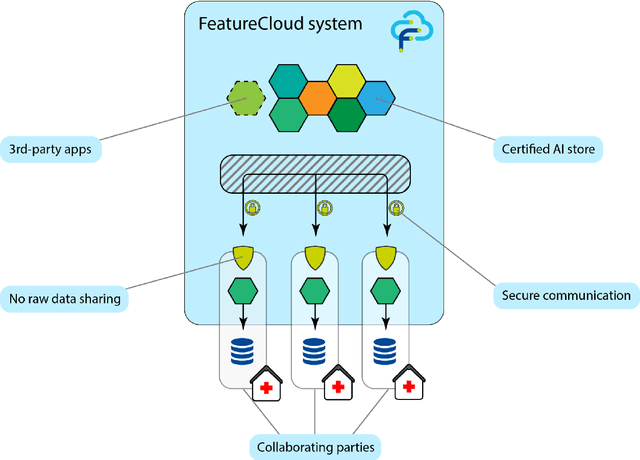
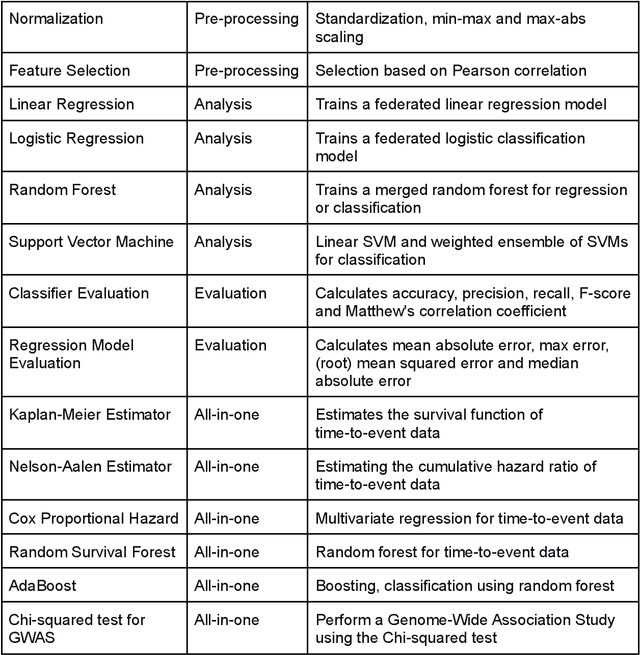
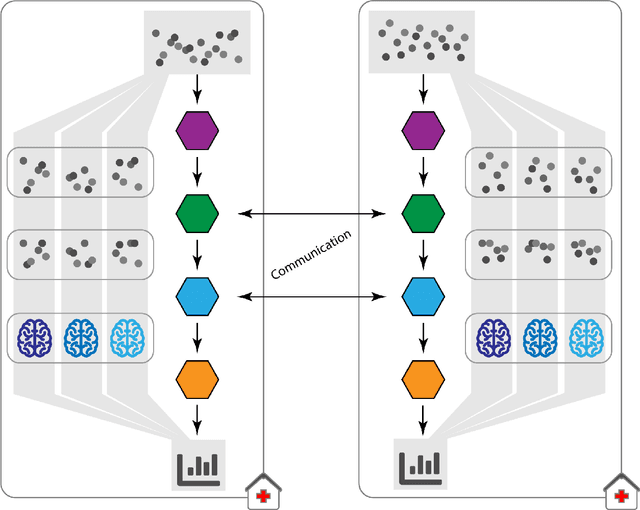
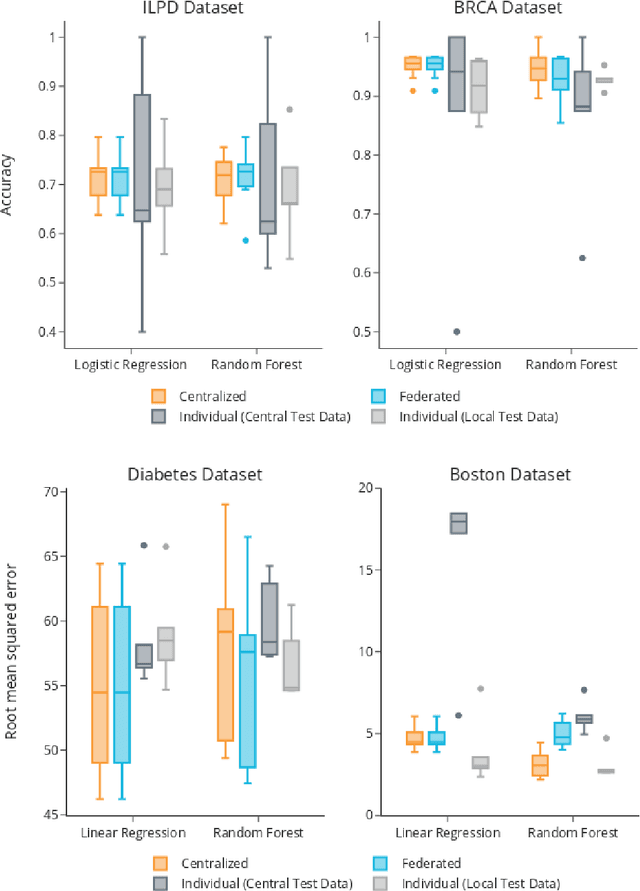
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
Federated Multi-Mini-Batch: An Efficient Training Approach to Federated Learning in Non-IID Environments
Nov 13, 2020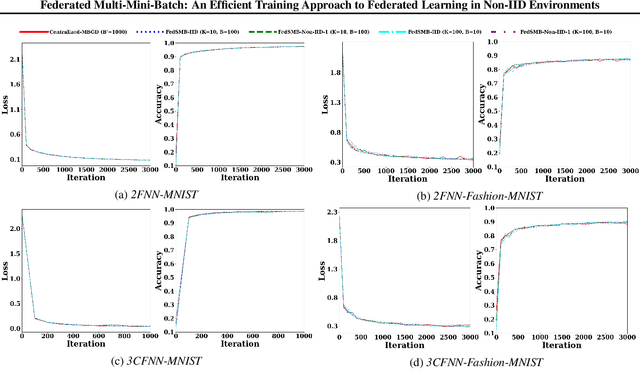
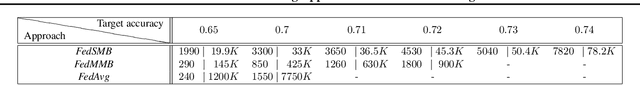
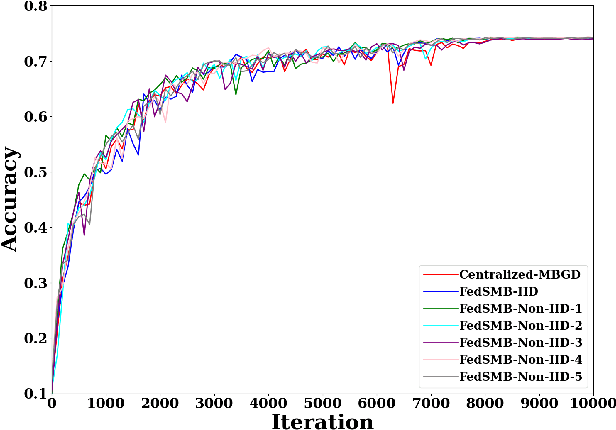
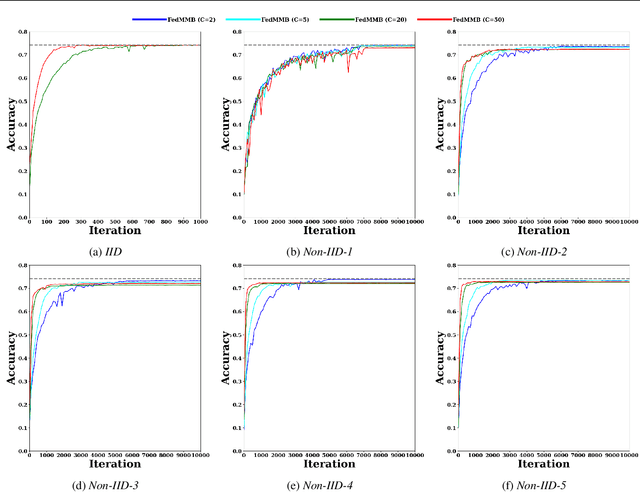
Abstract:Federated learning is a well-established approach to privacy-preserving training of a joint model on heavily distributed data. Federated averaging (FedAvg) is a well-known communication-efficient algorithm for federated learning, which performs well if the data distribution across the clients is independently and identically distributed (IID). However, FedAvg provides a lower accuracy and still requires a large number of communication rounds to achieve a target accuracy when it comes to Non-IID environments. To address the former limitation, we present federated single mini-batch (FedSMB), where the clients train the model on a single mini-batch from their dataset in each iteration. We show that FedSMB achieves the accuracy of the centralized training in Non-IID configurations, but in a considerable number of iterations. To address the latter limitation, we introduce federated multi-mini-batch (FedMMB) as a generalization of FedSMB, where the clients train the model on multiple mini-batches (specified by the batch count) in each communication round. FedMMB decouples the batch size from the batch count and provides a trade-off between the accuracy and communication efficiency in Non-IID settings. This is not possible with FedAvg, in which a single parameter determines both the batch size and batch count. The simulation results illustrate that FedMMB outperforms FedAvg in terms of the accuracy, communication efficiency, as well as computational efficiency and is an efficient training approach to federated learning in Non-IID environments.
Privacy-preserving Artificial Intelligence Techniques in Biomedicine
Jul 22, 2020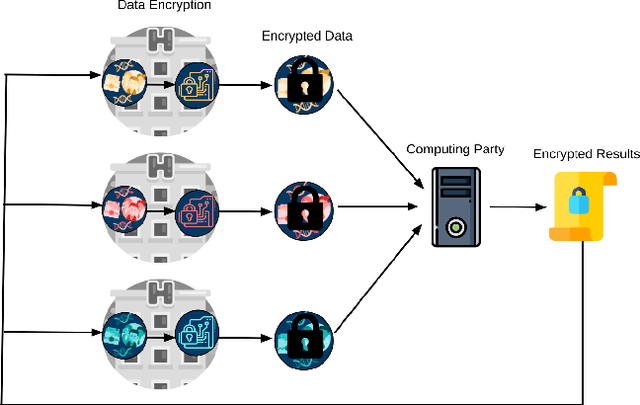
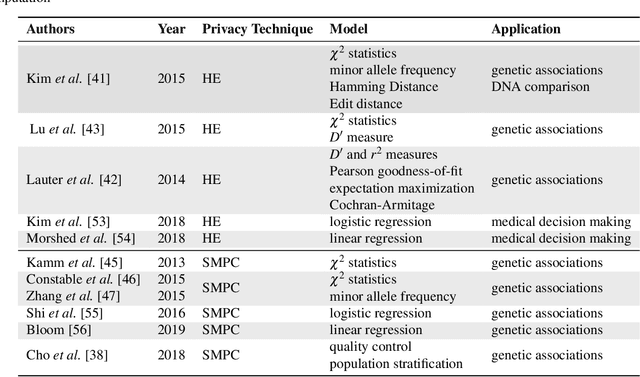
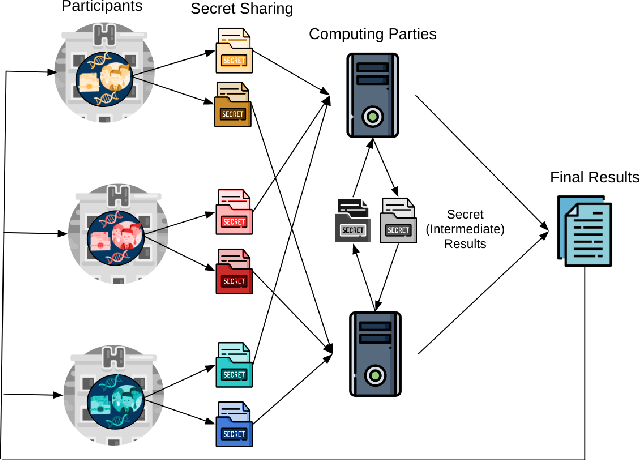
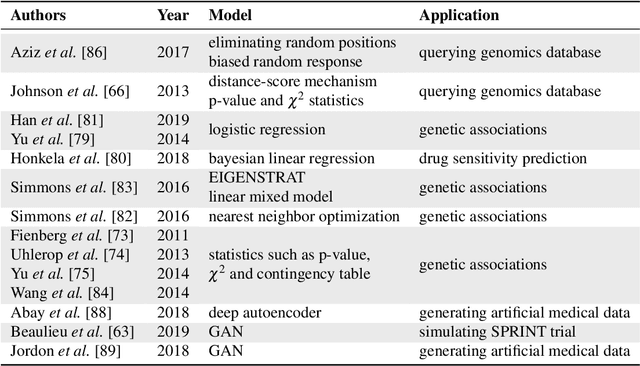
Abstract:Artificial intelligence (AI) has been successfully applied in numerous scientific domains including biomedicine and healthcare. Here, it has led to several breakthroughs ranging from clinical decision support systems, image analysis to whole genome sequencing. However, training an AI model on sensitive data raises also concerns about the privacy of individual participants. Adversary AIs, for example, can abuse even summary statistics of a study to determine the presence or absence of an individual in a given dataset. This has resulted in increasing restrictions to access biomedical data, which in turn is detrimental for collaborative research and impedes scientific progress. Hence there has been an explosive growth in efforts to harness the power of AI for learning from sensitive data while protecting patients' privacy. This paper provides a structured overview of recent advances in privacy-preserving AI techniques in biomedicine. It places the most important state-of-the-art approaches within a unified taxonomy, and discusses their strengths, limitations, and open problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge