Reza Nasirigerdeh
Improved Localized Machine Unlearning Through the Lens of Memorization
Dec 03, 2024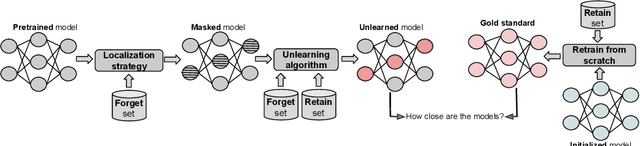
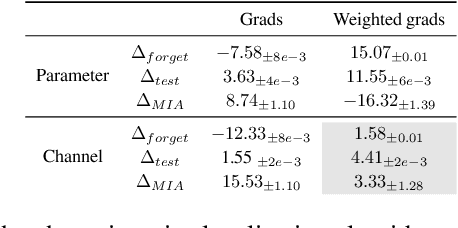
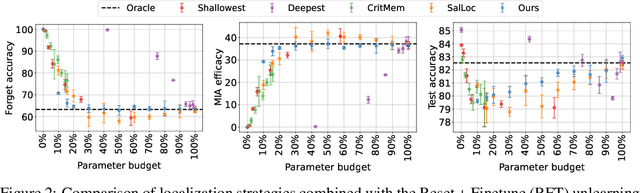
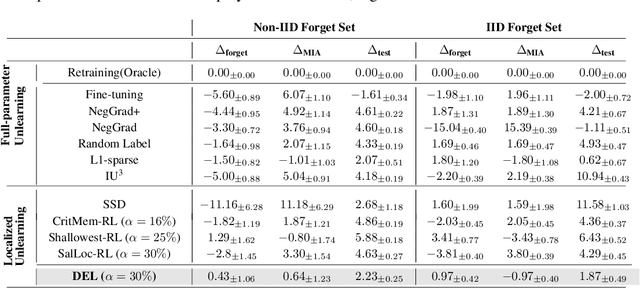
Abstract:Machine unlearning refers to removing the influence of a specified subset of training data from a machine learning model, efficiently, after it has already been trained. This is important for key applications, including making the model more accurate by removing outdated, mislabeled, or poisoned data. In this work, we study localized unlearning, where the unlearning algorithm operates on a (small) identified subset of parameters. Drawing inspiration from the memorization literature, we propose an improved localization strategy that yields strong results when paired with existing unlearning algorithms. We also propose a new unlearning algorithm, Deletion by Example Localization (DEL), that resets the parameters deemed-to-be most critical according to our localization strategy, and then finetunes them. Our extensive experiments on different datasets, forget sets and metrics reveal that DEL sets a new state-of-the-art for unlearning metrics, against both localized and full-parameter methods, while modifying a small subset of parameters, and outperforms the state-of-the-art localized unlearning in terms of test accuracy too.
Machine Unlearning for Medical Imaging
Jul 10, 2024

Abstract:Machine unlearning is the process of removing the impact of a particular set of training samples from a pretrained model. It aims to fulfill the "right to be forgotten", which grants the individuals such as patients the right to reconsider their contribution in models including medical imaging models. In this study, we evaluate the effectiveness (performance) and computational efficiency of different unlearning algorithms in medical imaging domain. Our evaluations demonstrate that the considered unlearning algorithms perform well on the retain set (samples whose influence on the model is allowed to be retained) and forget set (samples whose contribution to the model should be eliminated), and show no bias against male or female samples. They, however, adversely impact the generalization of the model, especially for larger forget set sizes. Moreover, they might be biased against easy or hard samples, and need additional computational overhead for hyper-parameter tuning. In conclusion, machine unlearning seems promising for medical imaging, but the existing unlearning algorithms still needs further improvements to become more practical for medical applications.
Label Noise-Robust Learning using a Confidence-Based Sieving Strategy
Oct 11, 2022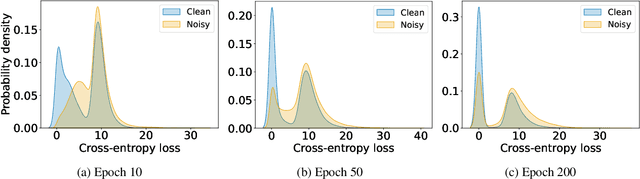
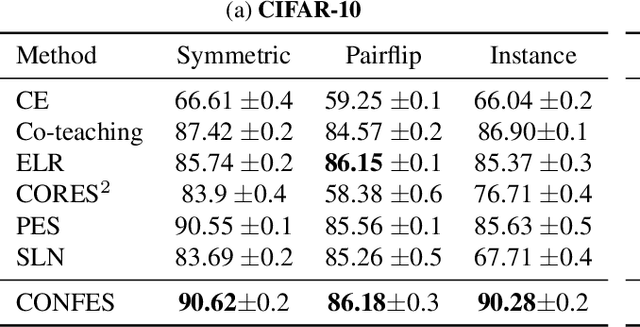
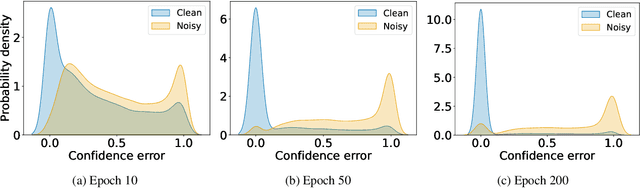
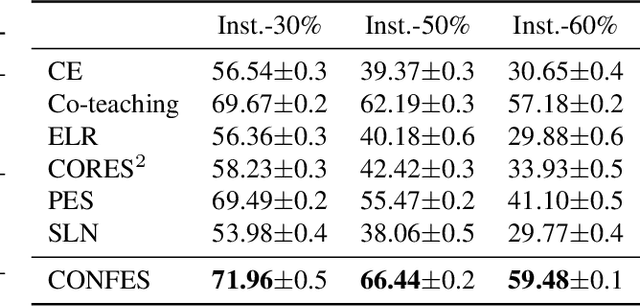
Abstract:In learning tasks with label noise, boosting model robustness against overfitting is a pivotal challenge because the model eventually memorizes labels including the noisy ones. Identifying the samples with corrupted labels and preventing the model from learning them is a promising approach to address this challenge. Per-sample training loss is a previously studied metric that considers samples with small loss as clean samples on which the model should be trained. In this work, we first demonstrate the ineffectiveness of this small-loss trick. Then, we propose a novel discriminator metric called confidence error and a sieving strategy called CONFES to effectively differentiate between the clean and noisy samples. We experimentally illustrate the superior performance of our proposed approach compared to recent studies on various settings such as synthetic and real-world label noise.
Kernel Normalized Convolutional Networks for Privacy-Preserving Machine Learning
Sep 30, 2022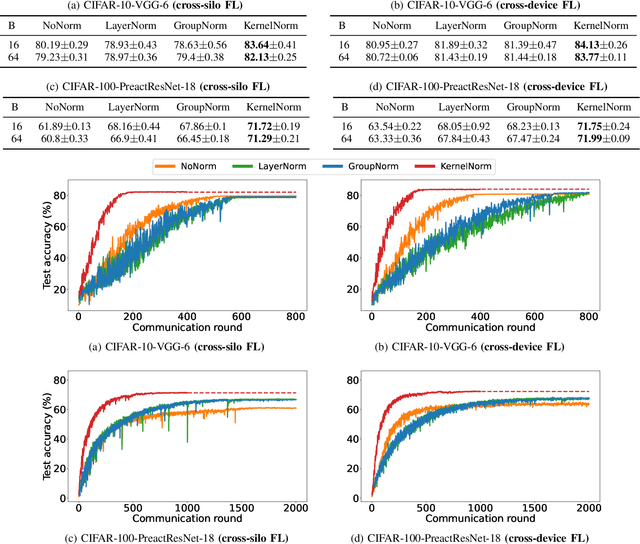
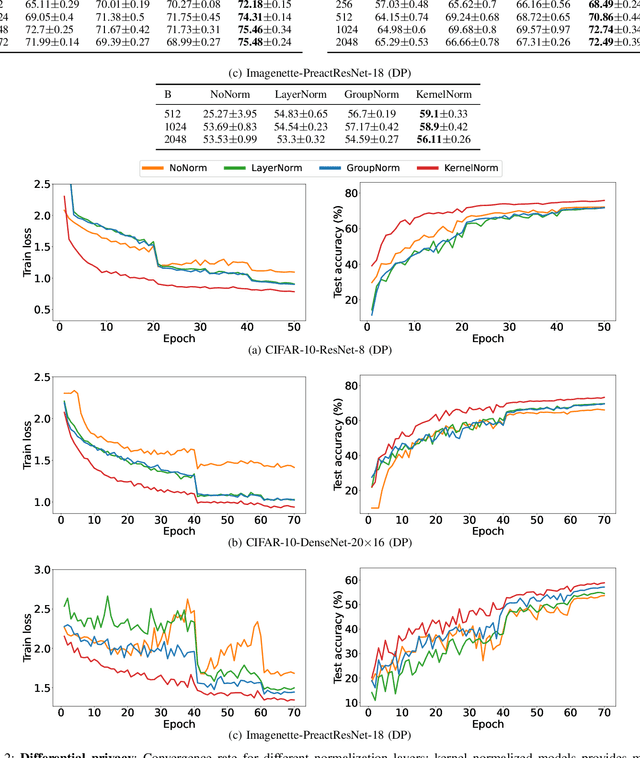
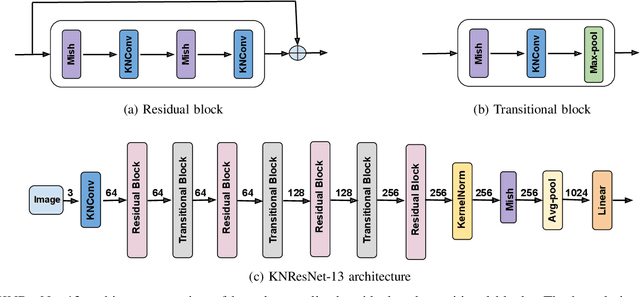

Abstract:Normalization is an important but understudied challenge in privacy-related application domains such as federated learning (FL) and differential privacy (DP). While the unsuitability of batch normalization for FL and DP has already been shown, the impact of the other normalization methods on the performance of federated or differentially private models is not well-known. To address this, we draw a performance comparison among layer normalization (LayerNorm), group normalization (GroupNorm), and the recently proposed kernel normalization (KernelNorm) in FL and DP settings. Our results indicate LayerNorm and GroupNorm provide no performance gain compared to the baseline (i.e. no normalization) for shallow models, but they considerably enhance performance of deeper models. KernelNorm, on the other hand, significantly outperforms its competitors in terms of accuracy and convergence rate (or communication efficiency) for both shallow and deeper models. Given these key observations, we propose a kernel normalized ResNet architecture called KNResNet-13 for differentially private learning environments. Using the proposed architecture, we provide new state-of-the-art accuracy values on the CIFAR-10 and Imagenette datasets.
Kernel Normalized Convolutional Networks
May 20, 2022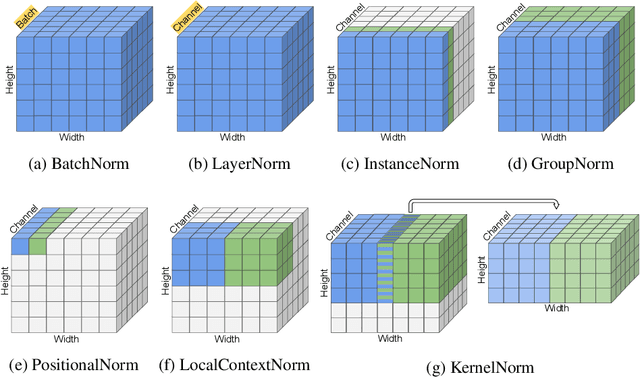
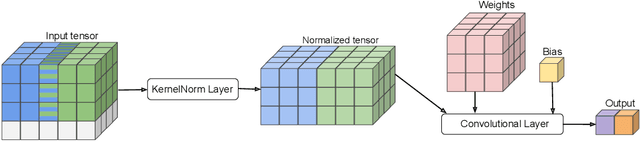

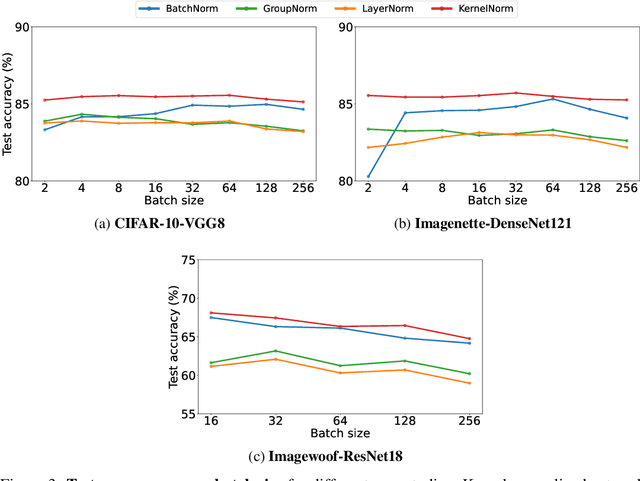
Abstract:Existing deep convolutional neural network (CNN) architectures frequently rely upon batch normalization (BatchNorm) to effectively train the model. BatchNorm significantly improves model performance, but performs poorly with smaller batch sizes. To address this limitation, we propose kernel normalization and kernel normalized convolutional layers, and incorporate them into kernel normalized convolutional networks (KNConvNets) as the main building blocks. We implement KNConvNets corresponding to the state-of-the-art CNNs such as ResNet and DenseNet while forgoing BatchNorm layers. Through extensive experiments, we illustrate that KNConvNets consistently outperform their batch, group, and layer normalized counterparts in terms of both accuracy and convergence rate while maintaining competitive computational efficiency.
HyFed: A Hybrid Federated Framework for Privacy-preserving Machine Learning
May 21, 2021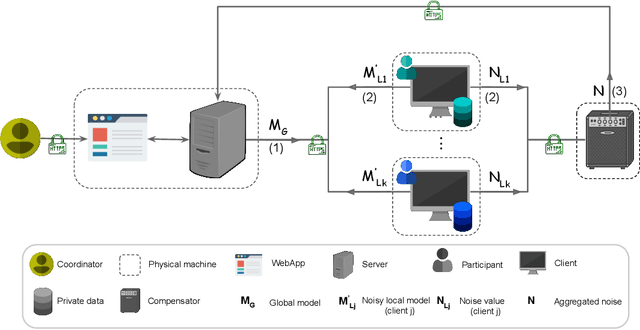

Abstract:Federated learning (FL) enables multiple clients to jointly train a global model under the coordination of a central server. Although FL is a privacy-aware paradigm, where raw data sharing is not required, recent studies have shown that FL might leak the private data of a client through the model parameters shared with the server or the other clients. In this paper, we present the HyFed framework, which enhances the privacy of FL while preserving the utility of the global model. HyFed provides developers with a generic API to develop federated, privacy-preserving algorithms. HyFed supports both simulation and federated operation modes and its source code is publicly available at https://github.com/tum-aimed/hyfed.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
May 12, 2021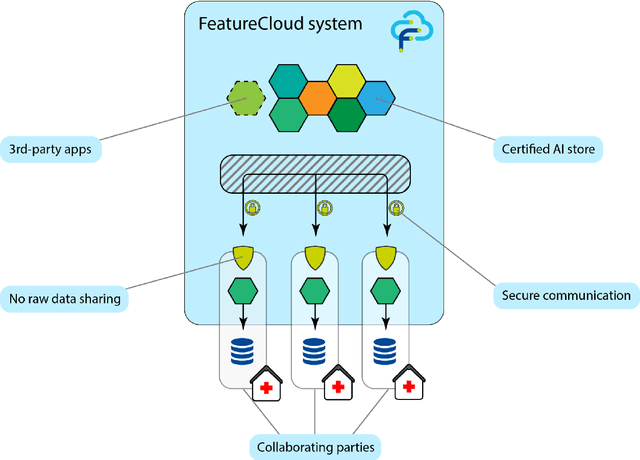
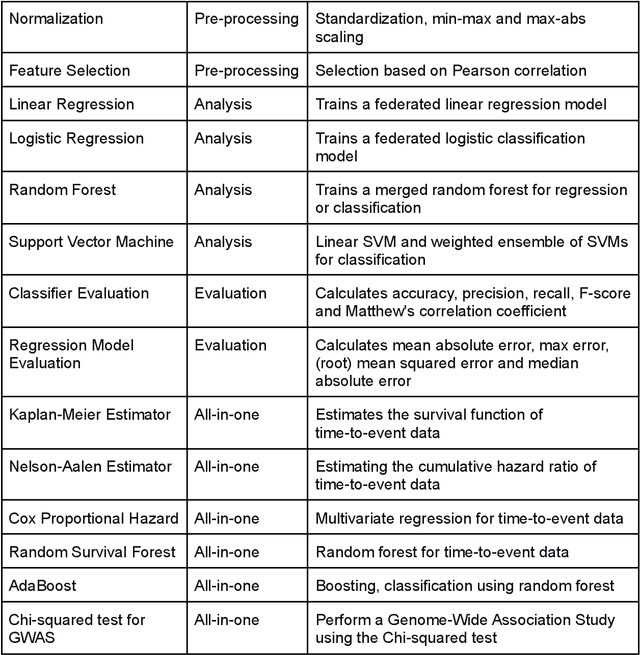
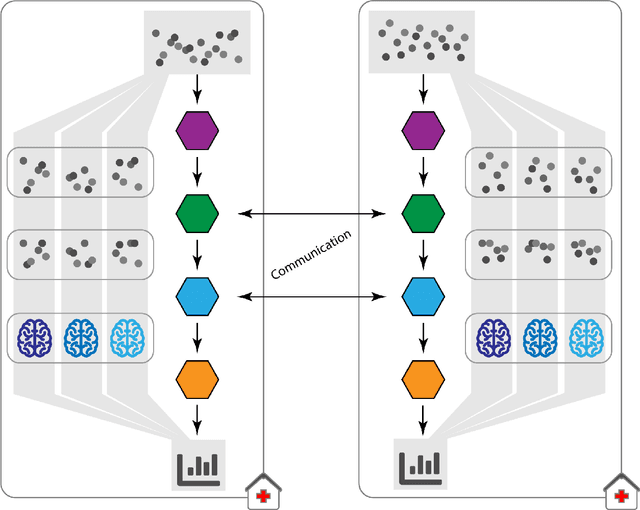
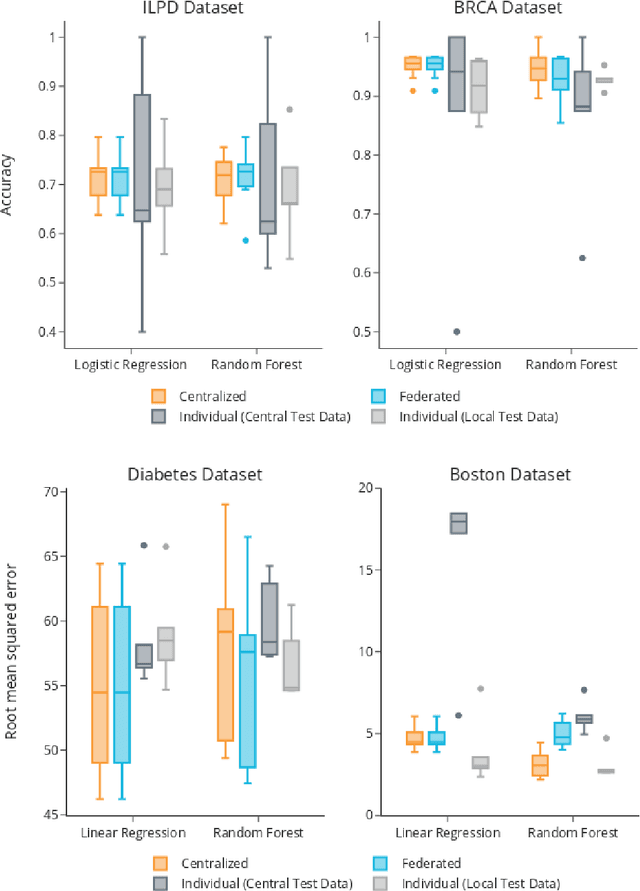
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
Federated Multi-Mini-Batch: An Efficient Training Approach to Federated Learning in Non-IID Environments
Nov 13, 2020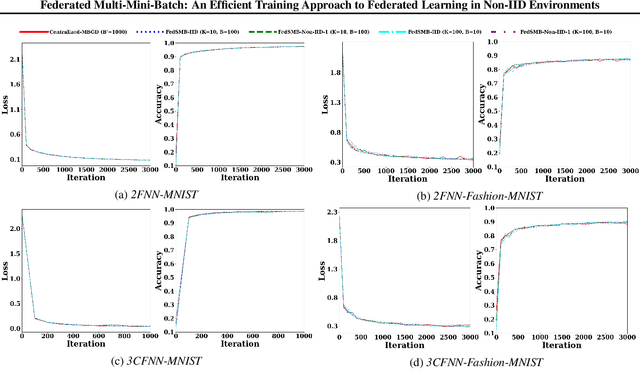
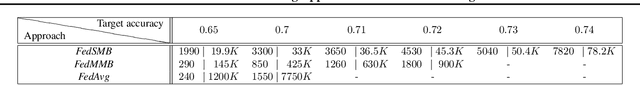
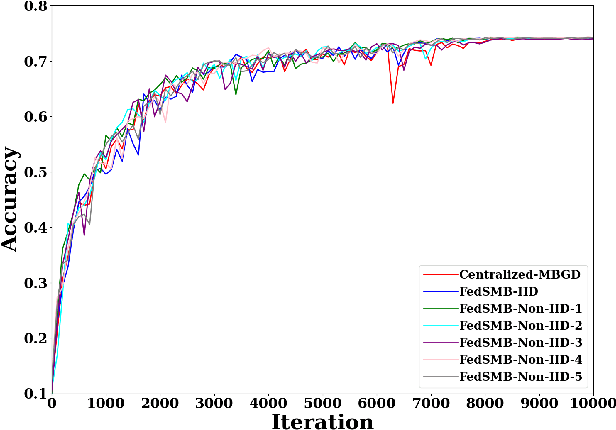
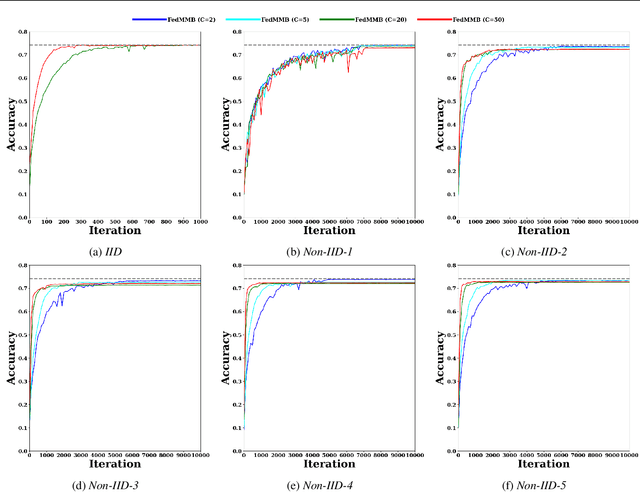
Abstract:Federated learning is a well-established approach to privacy-preserving training of a joint model on heavily distributed data. Federated averaging (FedAvg) is a well-known communication-efficient algorithm for federated learning, which performs well if the data distribution across the clients is independently and identically distributed (IID). However, FedAvg provides a lower accuracy and still requires a large number of communication rounds to achieve a target accuracy when it comes to Non-IID environments. To address the former limitation, we present federated single mini-batch (FedSMB), where the clients train the model on a single mini-batch from their dataset in each iteration. We show that FedSMB achieves the accuracy of the centralized training in Non-IID configurations, but in a considerable number of iterations. To address the latter limitation, we introduce federated multi-mini-batch (FedMMB) as a generalization of FedSMB, where the clients train the model on multiple mini-batches (specified by the batch count) in each communication round. FedMMB decouples the batch size from the batch count and provides a trade-off between the accuracy and communication efficiency in Non-IID settings. This is not possible with FedAvg, in which a single parameter determines both the batch size and batch count. The simulation results illustrate that FedMMB outperforms FedAvg in terms of the accuracy, communication efficiency, as well as computational efficiency and is an efficient training approach to federated learning in Non-IID environments.
Privacy-preserving Artificial Intelligence Techniques in Biomedicine
Jul 22, 2020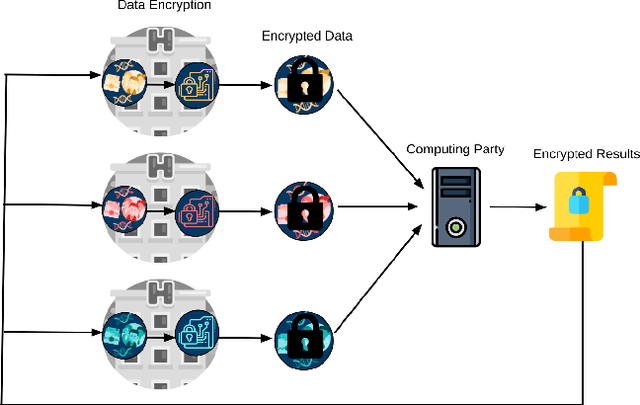
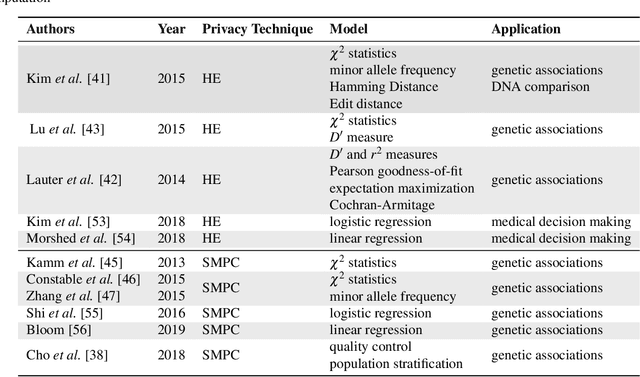
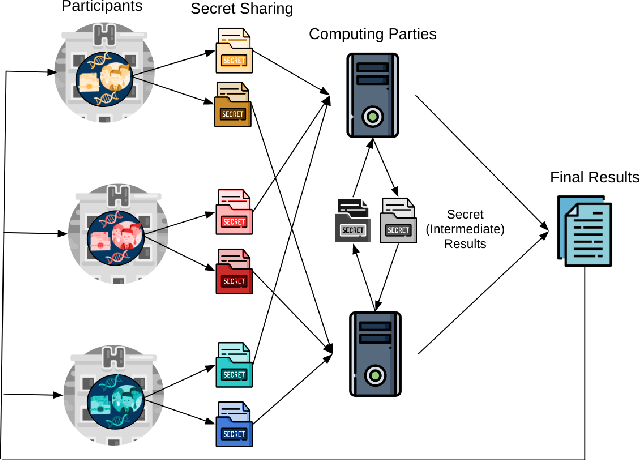
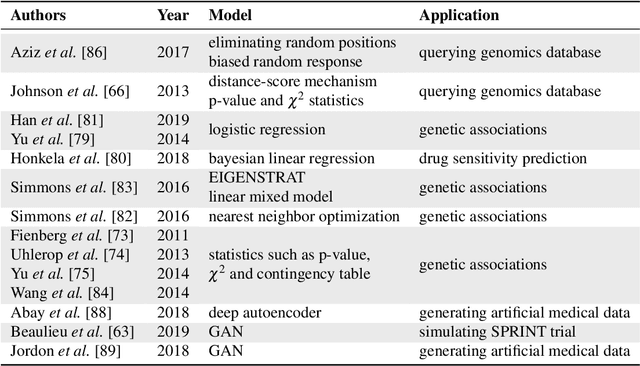
Abstract:Artificial intelligence (AI) has been successfully applied in numerous scientific domains including biomedicine and healthcare. Here, it has led to several breakthroughs ranging from clinical decision support systems, image analysis to whole genome sequencing. However, training an AI model on sensitive data raises also concerns about the privacy of individual participants. Adversary AIs, for example, can abuse even summary statistics of a study to determine the presence or absence of an individual in a given dataset. This has resulted in increasing restrictions to access biomedical data, which in turn is detrimental for collaborative research and impedes scientific progress. Hence there has been an explosive growth in efforts to harness the power of AI for learning from sensitive data while protecting patients' privacy. This paper provides a structured overview of recent advances in privacy-preserving AI techniques in biomedicine. It places the most important state-of-the-art approaches within a unified taxonomy, and discusses their strengths, limitations, and open problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge