Neil Lawrence
University of Sheffield
LLM Performance for Code Generation on Noisy Tasks
May 29, 2025Abstract:This paper investigates the ability of large language models (LLMs) to recognise and solve tasks which have been obfuscated beyond recognition. Focusing on competitive programming and benchmark tasks (LeetCode and MATH), we compare performance across multiple models and obfuscation methods, such as noise and redaction. We demonstrate that all evaluated LLMs can solve tasks obfuscated to a level where the text would be unintelligible to human readers, and does not contain key pieces of instruction or context. We introduce the concept of eager pattern matching to describe this behaviour, which is not observed in tasks published after the models' knowledge cutoff date, indicating strong memorisation or overfitting to training data, rather than legitimate reasoning about the presented problem. We report empirical evidence of distinct performance decay patterns between contaminated and unseen datasets. We discuss the implications for benchmarking and evaluations of model behaviour, arguing for caution when designing experiments using standard datasets. We also propose measuring the decay of performance under obfuscation as a possible strategy for detecting dataset contamination and highlighting potential safety risks and interpretability issues for automated software systems.
Meta-Surrogate Benchmarking for Hyperparameter Optimization
May 30, 2019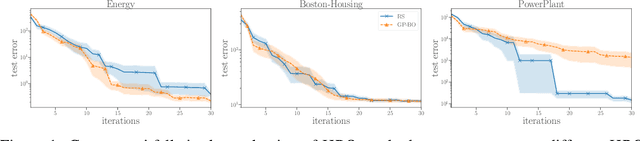
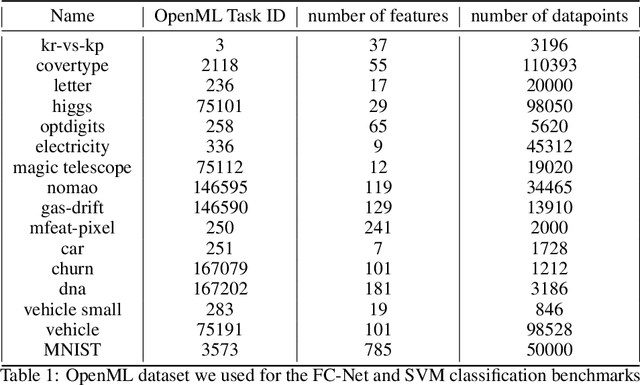
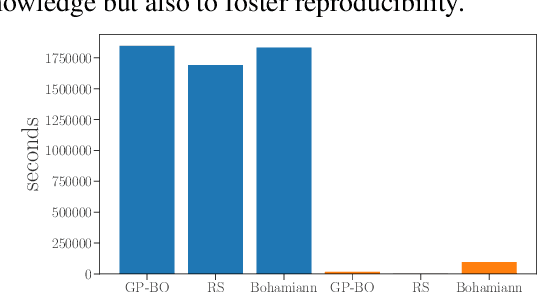
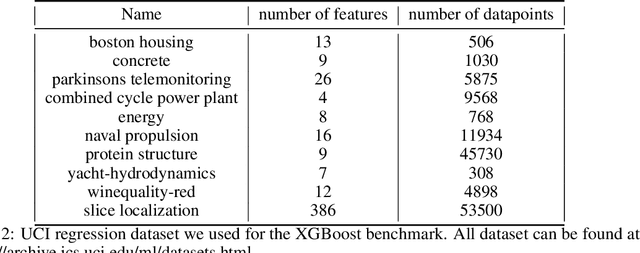
Abstract:Despite the recent progress in hyperparameter optimization (HPO), available benchmarks that resemble real-world scenarios consist of a few and very large problem instances that are expensive to solve. This blocks researchers and practitioners not only from systematically running large-scale comparisons that are needed to draw statistically significant results but also from reproducing experiments that were conducted before. This work proposes a method to alleviate these issues by means of a meta-surrogate model for HPO tasks trained on off-line generated data. The model combines a probabilistic encoder with a multi-task model such that it can generate inexpensive and realistic tasks of the class of problems of interest. We demonstrate that benchmarking HPO methods on samples of the generative model allows us to draw more coherent and statistically significant conclusions that can be reached orders of magnitude faster than using the original tasks. We provide evidence of our findings for various HPO methods on a wide class of problems.
Facilitating Bayesian Continual Learning by Natural Gradients and Stein Gradients
Apr 24, 2019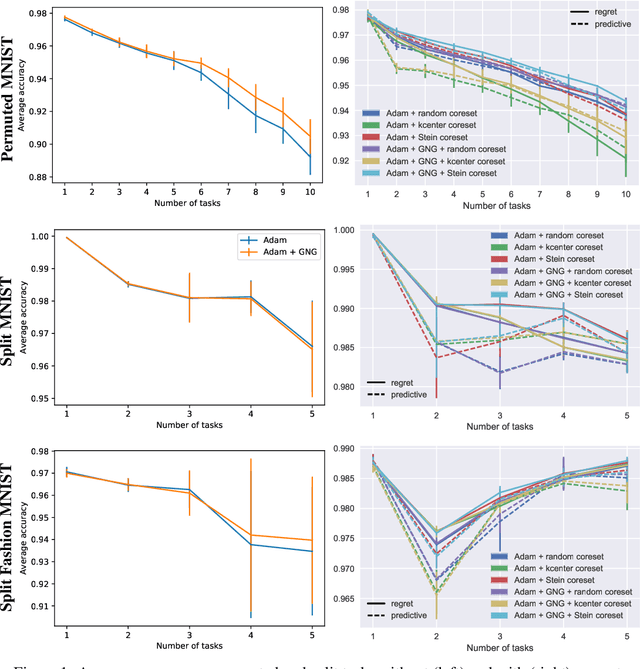
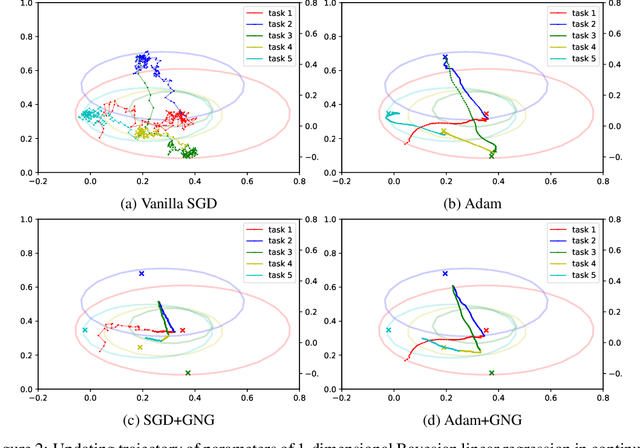
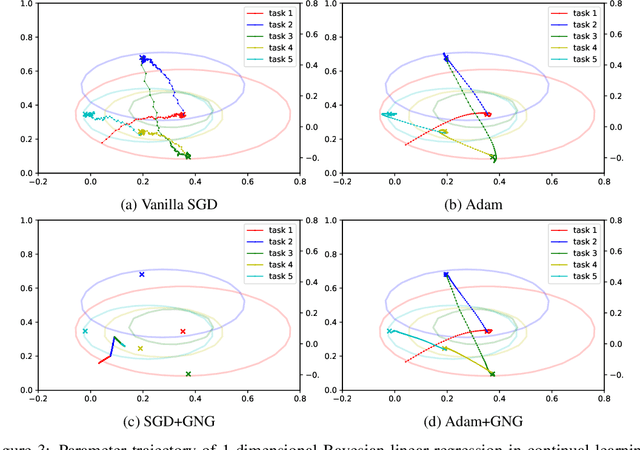
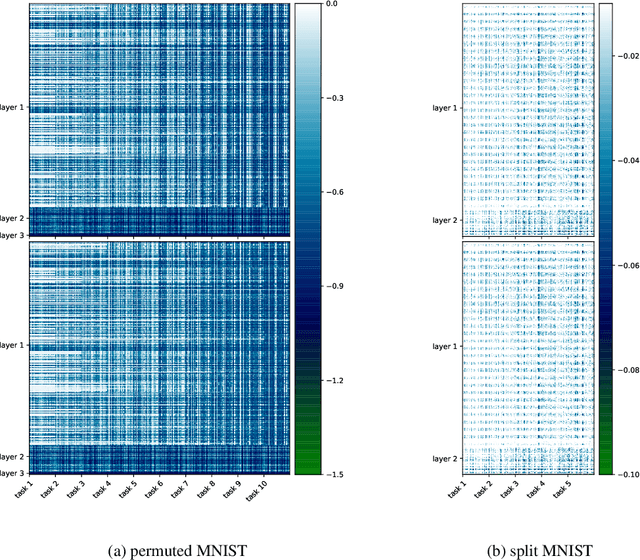
Abstract:Continual learning aims to enable machine learning models to learn a general solution space for past and future tasks in a sequential manner. Conventional models tend to forget the knowledge of previous tasks while learning a new task, a phenomenon known as catastrophic forgetting. When using Bayesian models in continual learning, knowledge from previous tasks can be retained in two ways: 1). posterior distributions over the parameters, containing the knowledge gained from inference in previous tasks, which then serve as the priors for the following task; 2). coresets, containing knowledge of data distributions of previous tasks. Here, we show that Bayesian continual learning can be facilitated in terms of these two means through the use of natural gradients and Stein gradients respectively.
Continual Learning in Practice
Mar 18, 2019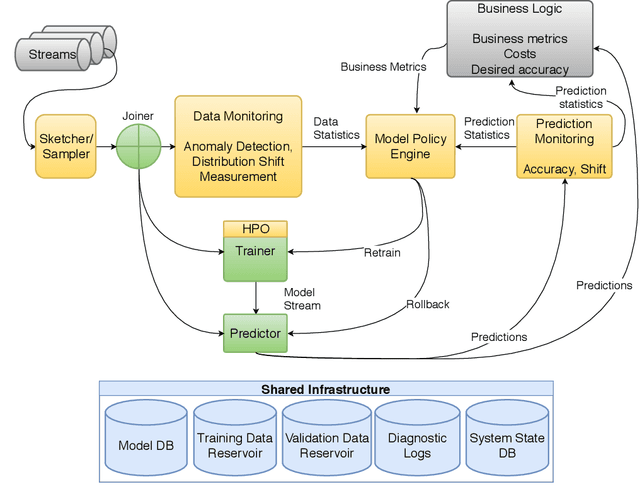
Abstract:This paper describes a reference architecture for self-maintaining systems that can learn continually, as data arrives. In environments where data evolves, we need architectures that manage Machine Learning (ML) models in production, adapt to shifting data distributions, cope with outliers, retrain when necessary, and adapt to new tasks. This represents continual AutoML or Automatically Adaptive Machine Learning. We describe the challenges and proposes a reference architecture.
Deep Gaussian Processes for Multi-fidelity Modeling
Mar 18, 2019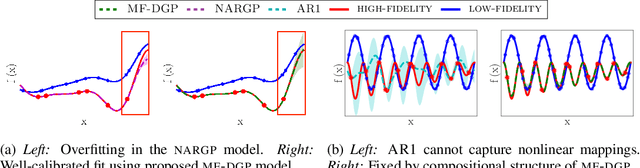
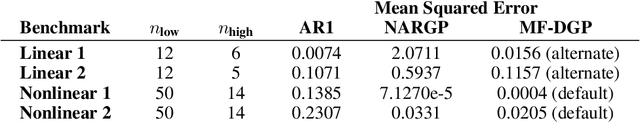
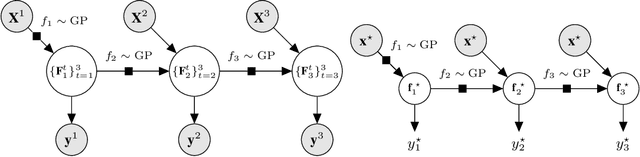
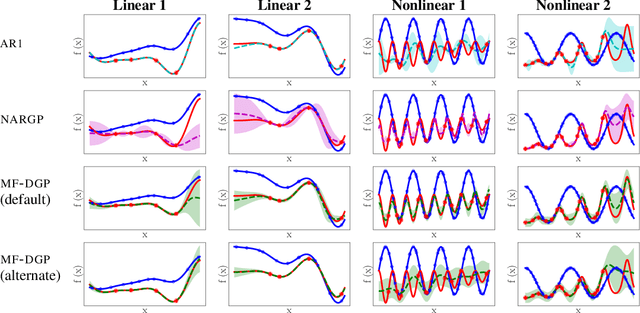
Abstract:Multi-fidelity methods are prominently used when cheaply-obtained, but possibly biased and noisy, observations must be effectively combined with limited or expensive true data in order to construct reliable models. This arises in both fundamental machine learning procedures such as Bayesian optimization, as well as more practical science and engineering applications. In this paper we develop a novel multi-fidelity model which treats layers of a deep Gaussian process as fidelity levels, and uses a variational inference scheme to propagate uncertainty across them. This allows for capturing nonlinear correlations between fidelities with lower risk of overfitting than existing methods exploiting compositional structure, which are conversely burdened by structural assumptions and constraints. We show that the proposed approach makes substantial improvements in quantifying and propagating uncertainty in multi-fidelity set-ups, which in turn improves their effectiveness in decision making pipelines.
Intrinsic Gaussian processes on complex constrained domains
Jan 03, 2018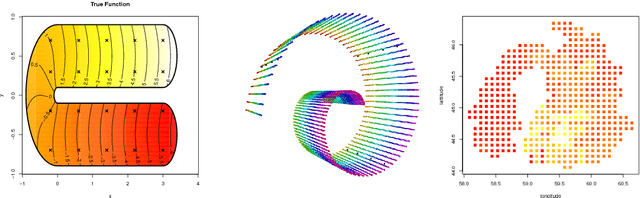
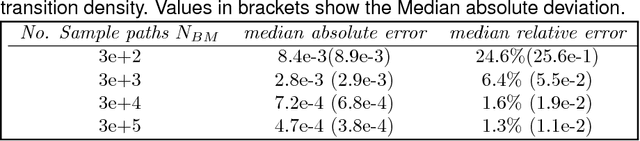
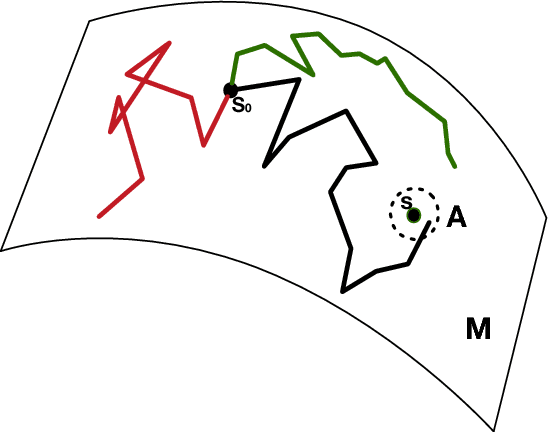

Abstract:We propose a class of intrinsic Gaussian processes (in-GPs) for interpolation, regression and classification on manifolds with a primary focus on complex constrained domains or irregular shaped spaces arising as subsets or submanifolds of R, R2, R3 and beyond. For example, in-GPs can accommodate spatial domains arising as complex subsets of Euclidean space. in-GPs respect the potentially complex boundary or interior conditions as well as the intrinsic geometry of the spaces. The key novelty of the proposed approach is to utilise the relationship between heat kernels and the transition density of Brownian motion on manifolds for constructing and approximating valid and computationally feasible covariance kernels. This enables in-GPs to be practically applied in great generality, while existing approaches for smoothing on constrained domains are limited to simple special cases. The broad utilities of the in-GP approach is illustrated through simulation studies and data examples.
Parallelizable sparse inverse formulation Gaussian processes (SpInGP)
Sep 28, 2017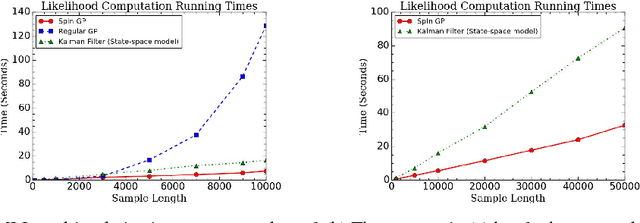
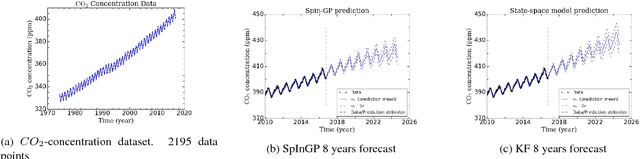
Abstract:We propose a parallelizable sparse inverse formulation Gaussian process (SpInGP) for temporal models. It uses a sparse precision GP formulation and sparse matrix routines to speed up the computations. Due to the state-space formulation used in the algorithm, the time complexity of the basic SpInGP is linear, and because all the computations are parallelizable, the parallel form of the algorithm is sublinear in the number of data points. We provide example algorithms to implement the sparse matrix routines and experimentally test the method using both simulated and real data.
Spatio-temporal Gaussian processes modeling of dynamical systems in systems biology
Oct 17, 2016
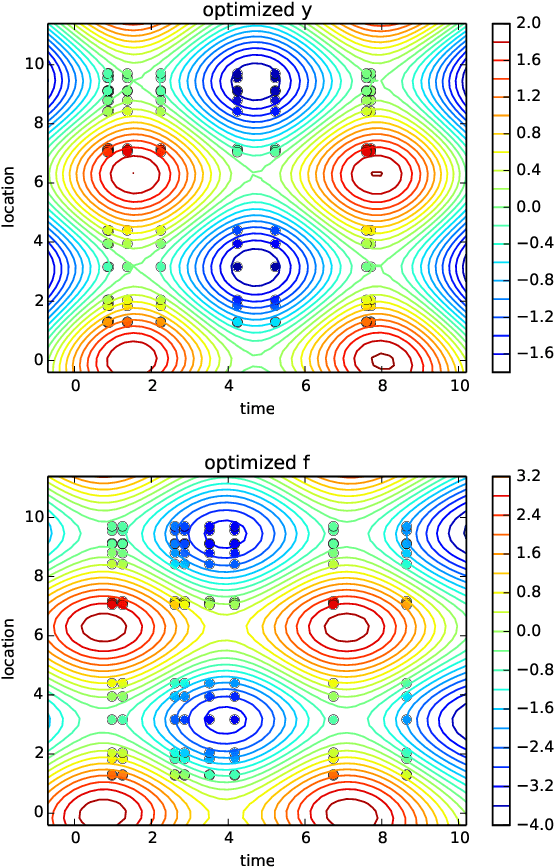
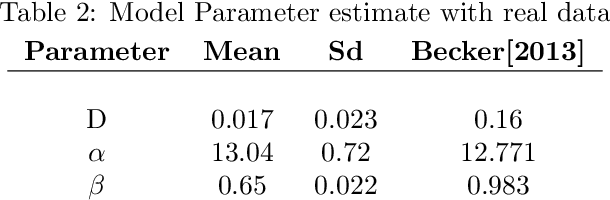
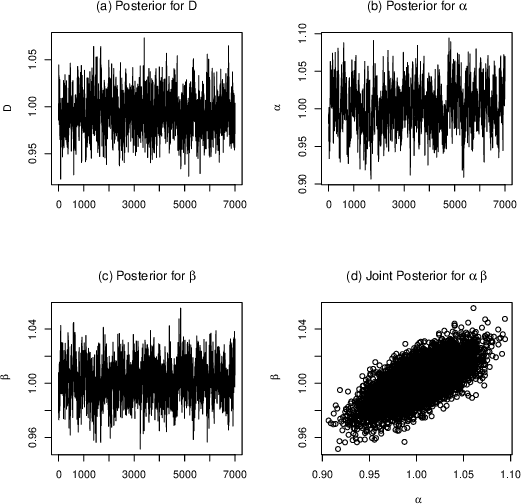
Abstract:Quantitative modeling of post-transcriptional regulation process is a challenging problem in systems biology. A mechanical model of the regulatory process needs to be able to describe the available spatio-temporal protein concentration and mRNA expression data and recover the continuous spatio-temporal fields. Rigorous methods are required to identify model parameters. A promising approach to deal with these difficulties is proposed using Gaussian process as a prior distribution over the latent function of protein concentration and mRNA expression. In this study, we consider a partial differential equation mechanical model with differential operators and latent function. Since the operators at stake are linear, the information from the physical model can be encoded into the kernel function. Hybrid Monte Carlo methods are employed to carry out Bayesian inference of the partial differential equation parameters and Gaussian process kernel parameters. The spatio-temporal field of protein concentration and mRNA expression are reconstructed without explicitly solving the partial differential equation.
Unsupervised Learning with Imbalanced Data via Structure Consolidation Latent Variable Model
Jun 30, 2016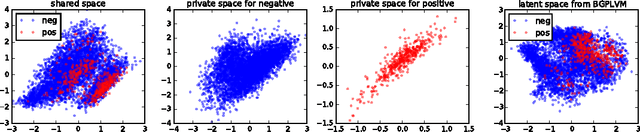
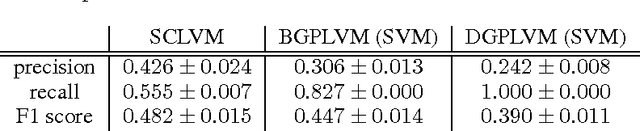
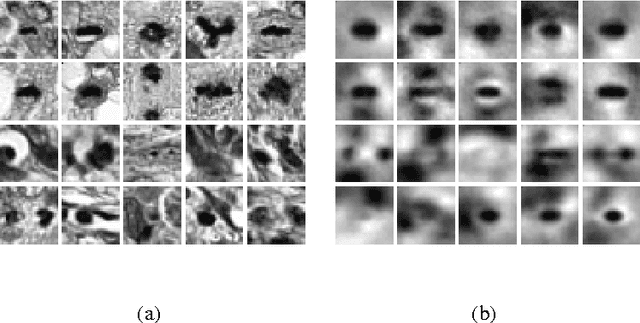
Abstract:Unsupervised learning on imbalanced data is challenging because, when given imbalanced data, current model is often dominated by the major category and ignores the categories with small amount of data. We develop a latent variable model that can cope with imbalanced data by dividing the latent space into a shared space and a private space. Based on Gaussian Process Latent Variable Models, we propose a new kernel formulation that enables the separation of latent space and derives an efficient variational inference method. The performance of our model is demonstrated with an imbalanced medical image dataset.
Variational Auto-encoded Deep Gaussian Processes
Feb 29, 2016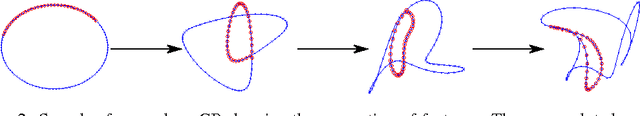
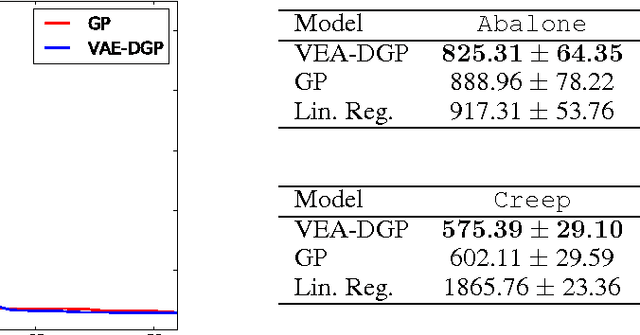
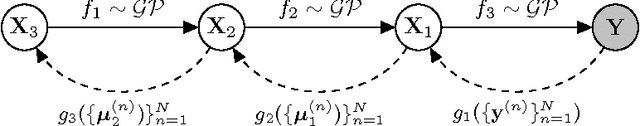
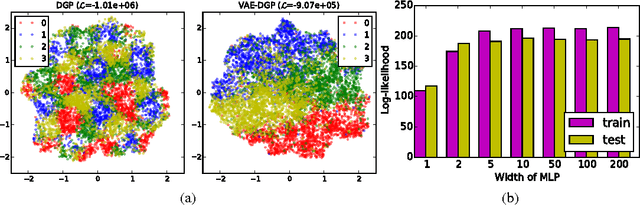
Abstract:We develop a scalable deep non-parametric generative model by augmenting deep Gaussian processes with a recognition model. Inference is performed in a novel scalable variational framework where the variational posterior distributions are reparametrized through a multilayer perceptron. The key aspect of this reformulation is that it prevents the proliferation of variational parameters which otherwise grow linearly in proportion to the sample size. We derive a new formulation of the variational lower bound that allows us to distribute most of the computation in a way that enables to handle datasets of the size of mainstream deep learning tasks. We show the efficacy of the method on a variety of challenges including deep unsupervised learning and deep Bayesian optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge