Yeman Brhane Hagos
MiceBoneChallenge: Micro-CT public dataset and six solutions for automatic growth plate detection in micro-CT mice bone scans
Nov 26, 2024



Abstract:Detecting and quantifying bone changes in micro-CT scans of rodents is a common task in preclinical drug development studies. However, this task is manual, time-consuming and subject to inter- and intra-observer variability. In 2024, Anonymous Company organized an internal challenge to develop models for automatic bone quantification. We prepared and annotated a high-quality dataset of 3D $\mu$CT bone scans from $83$ mice. The challenge attracted over $80$ AI scientists from around the globe who formed $23$ teams. The participants were tasked with developing a solution to identify the plane where the bone growth happens, which is essential for fully automatic segmentation of trabecular bone. As a result, six computer vision solutions were developed that can accurately identify the location of the growth plate plane. The solutions achieved the mean absolute error of $1.91\pm0.87$ planes from the ground truth on the test set, an accuracy level acceptable for practical use by a radiologist. The annotated 3D scans dataset along with the six solutions and source code, is being made public, providing researchers with opportunities to develop and benchmark their own approaches. The code, trained models, and the data will be shared.
Lung tumor segmentation in MRI mice scans using 3D nnU-Net with minimum annotations
Nov 01, 2024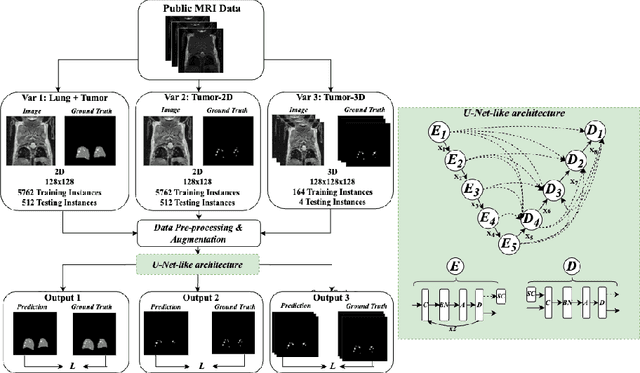
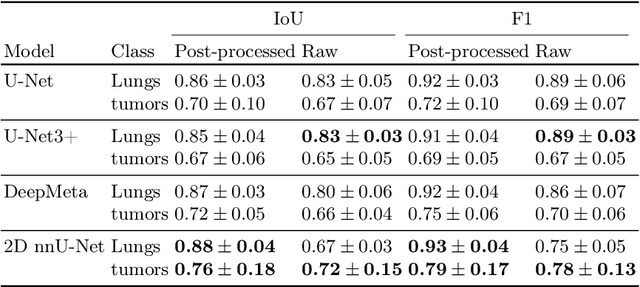
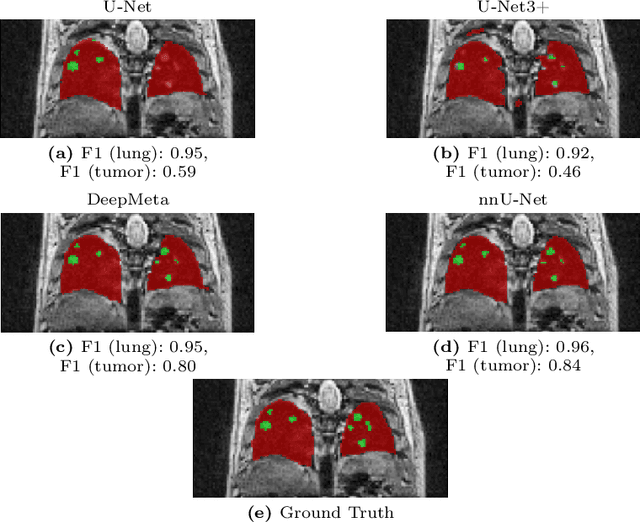
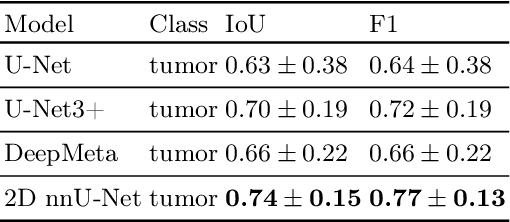
Abstract:In drug discovery, accurate lung tumor segmentation is an important step for assessing tumor size and its progression using \textit{in-vivo} imaging such as MRI. While deep learning models have been developed to automate this process, the focus has predominantly been on human subjects, neglecting the pivotal role of animal models in pre-clinical drug development. In this work, we focus on optimizing lung tumor segmentation in mice. First, we demonstrate that the nnU-Net model outperforms the U-Net, U-Net3+, and DeepMeta models. Most importantly, we achieve better results with nnU-Net 3D models than 2D models, indicating the importance of spatial context for segmentation tasks in MRI mice scans. This study demonstrates the importance of 3D input over 2D input images for lung tumor segmentation in MRI scans. Finally, we outperform the prior state-of-the-art approach that involves the combined segmentation of lungs and tumors within the lungs. Our work achieves comparable results using only lung tumor annotations requiring fewer annotations, saving time and annotation efforts. This work\footnote{\url{https://anonymous.4open.science/r/lung-tumour-mice-mri-64BB}} is an important step in automating pre-clinical animal studies to quantify the efficacy of experimental drugs, particularly in assessing tumor changes.
Cell abundance aware deep learning for cell detection on highly imbalanced pathological data
Feb 23, 2021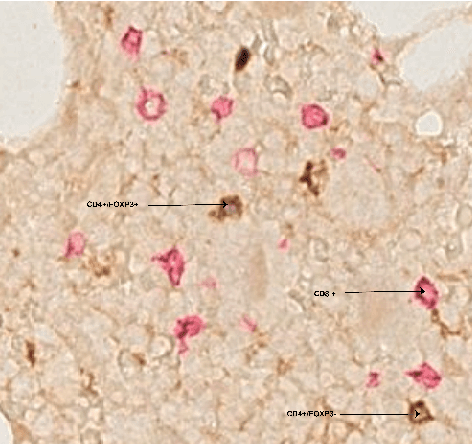


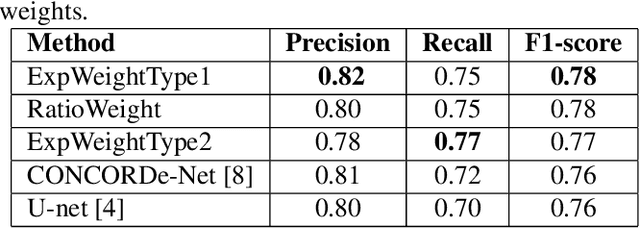
Abstract:Automated analysis of tissue sections allows a better understanding of disease biology and may reveal biomarkers that could guide prognosis or treatment selection. In digital pathology, less abundant cell types can be of biological significance, but their scarcity can result in biased and sub-optimal cell detection model. To minimize the effect of cell imbalance on cell detection, we proposed a deep learning pipeline that considers the abundance of cell types during model training. Cell weight images were generated, which assign larger weights to less abundant cells and used the weights to regularize Dice overlap loss function. The model was trained and evaluated on myeloma bone marrow trephine samples. Our model obtained a cell detection F1-score of 0.78, a 2% increase compared to baseline models, and it outperformed baseline models at detecting rare cell types. We found that scaling deep learning loss function by the abundance of cells improves cell detection performance. Our results demonstrate the importance of incorporating domain knowledge on deep learning methods for pathological data with class imbalance.
ConCORDe-Net: Cell Count Regularized Convolutional Neural Network for Cell Detection in Multiplex Immunohistochemistry Images
Aug 01, 2019
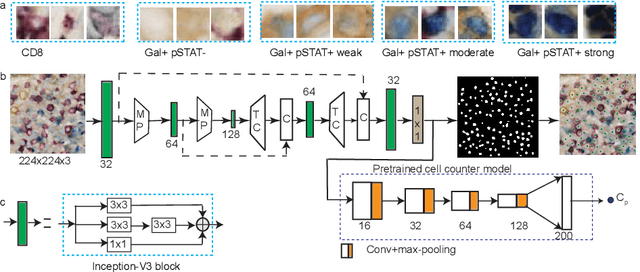

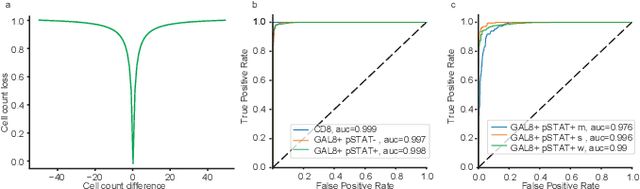
Abstract:In digital pathology, cell detection and classification are often prerequisites to quantify cell abundance and explore tissue spatial heterogeneity. However, these tasks are particularly challenging for multiplex immunohistochemistry (mIHC) images due to high levels of variability in staining, expression intensity, and inherent noise as a result of preprocessing artefacts. We proposed a deep learning method to detect and classify cells in mIHC whole-tumour slide images of breast cancer. Inspired by inception-v3, we developed Cell COunt RegularizeD Convolutional neural Network (ConCORDe-Net) which integrates conventional dice overlap and a new cell count loss function for optimizing cell detection, followed by a multi-stage convolutional neural network for cell classification. In total, 20447 cells, belonging to five cell classes were annotated by experts from 175 patches extracted from 6 whole-tumour mIHC images. These patches were randomly split into training, validation and testing sets. Using ConCORDe-Net, we obtained a cell detection F1 score of 0.873, which is the best score compared to three state of the art methods. In particular, ConCORDe-Net excels at detecting closely located and weakly stained cells compared to other methods. Incorporating cell count loss in the objective function regularizes the network to learn weak gradient boundaries and separate weakly stained cells from background artefacts. Moreover, cell classification accuracy of 96.5% was achieved. These results support that incorporating problem-specific knowledge such as cell count into deep learning-based cell detection architectures improve the robustness of the algorithm.
Improving Breast Cancer Detection using Symmetry Information with Deep Learning
Aug 17, 2018



Abstract:Convolutional Neural Networks (CNN) have had a huge success in many areas of computer vision and medical image analysis. However, there is still an immense potential for performance improvement in mammogram breast cancer detection Computer-Aided Detection (CAD) systems by integrating all the information that the radiologist utilizes, such as symmetry and temporal data. In this work, we proposed a patch based multi-input CNN that learns symmetrical difference to detect breast masses. The network was trained on a large-scale dataset of 28294 mammogram images. The performance was compared to a baseline architecture without symmetry context using Area Under the ROC Curve (AUC) and Competition Performance Metric (CPM). At candidate level, AUC value of 0.933 with 95% confidence interval of [0.920, 0.954] was obtained when symmetry information is incorporated in comparison with baseline architecture which yielded AUC value of 0.929 with [0.919, 0.947] confidence interval. By incorporating symmetrical information, although there was no a significant candidate level performance again (p = 0.111), we have found a compelling result at exam level with CPM value of 0.733 (p = 0.001). We believe that including temporal data, and adding benign class to the dataset could improve the detection performance.
Smoothness-based Edge Detection using Low-SNR Camera for Robot Navigation
Oct 03, 2017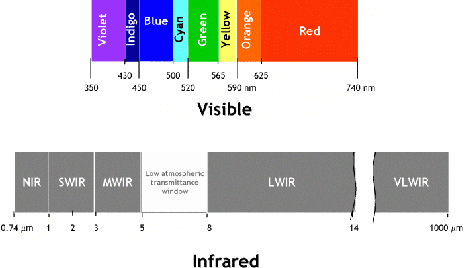
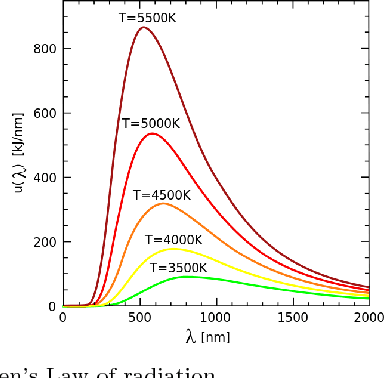
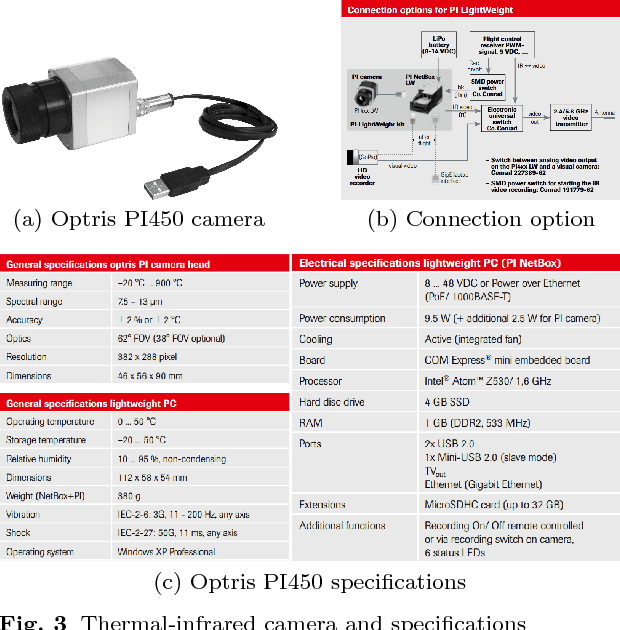

Abstract:In the emerging advancement in the branch of autonomous robotics, the ability of a robot to efficiently localize and construct maps of its surrounding is crucial. This paper deals with utilizing thermal-infrared cameras, as opposed to conventional cameras as the primary sensor to capture images of the robot's surroundings. For localization, the images need to be further processed before feeding them to a navigational system. The main motivation of this paper was to develop an edge detection methodology capable of utilizing the low-SNR poor output from such a thermal camera and effectively detect smooth edges of the surrounding environment. The enhanced edge detector proposed in this paper takes the raw image from the thermal sensor, denoises the images, applies Canny edge detection followed by CSS method. The edges are ranked to remove any noise and only edges of the highest rank are kept. Then, the broken edges are linked by computing edge metrics and a smooth edge of the surrounding is displayed in a binary image. Several comparisons are also made in the paper between the proposed technique and the existing techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge