Fahdi Kanavati
$σ$-PCA: a unified neural model for linear and nonlinear principal component analysis
Nov 25, 2023



Abstract:Linear principal component analysis (PCA), nonlinear PCA, and linear independent component analysis (ICA) -- those are three methods with single-layer autoencoder formulations for learning linear transformations from data. Linear PCA learns orthogonal transformations (rotations) that orient axes to maximise variance, but it suffers from a subspace rotational indeterminacy: it fails to find a unique rotation for axes that share the same variance. Both nonlinear PCA and linear ICA reduce the subspace indeterminacy from rotational to permutational by maximising statistical independence under the assumption of unit variance. The relationship between all three can be understood by the singular value decomposition of the linear ICA transformation into a sequence of rotation, scale, rotation. Linear PCA learns the first rotation; nonlinear PCA learns the second. The scale is simply the inverse of the standard deviations. The problem is that, in contrast to linear PCA, conventional nonlinear PCA cannot be used directly on the data to learn the first rotation, the first being special as it reduces dimensionality and orders by variances. In this paper, we have identified the cause, and as a solution we propose $\sigma$-PCA: a unified neural model for linear and nonlinear PCA as single-layer autoencoders. One of its key ingredients: modelling not just the rotation but also the scale -- the variances. This model bridges the disparity between linear and nonlinear PCA. And so, like linear PCA, it can learn a semi-orthogonal transformation that reduces dimensionality and orders by variances, but, unlike linear PCA, it does not suffer from rotational indeterminacy.
Inference of captions from histopathological patches
Feb 07, 2022



Abstract:Computational histopathology has made significant strides in the past few years, slowly getting closer to clinical adoption. One area of benefit would be the automatic generation of diagnostic reports from H\&E-stained whole slide images which would further increase the efficiency of the pathologists' routine diagnostic workflows. In this study, we compiled a dataset (PatchGastricADC22) of histopathological captions of stomach adenocarcinoma endoscopic biopsy specimens, which we extracted from diagnostic reports and paired with patches extracted from the associated whole slide images. The dataset contains a variety of gastric adenocarcinoma subtypes. We trained a baseline attention-based model to predict the captions from features extracted from the patches and obtained promising results. We make the captioned dataset of 262K patches publicly available.
Bayesian analysis of the prevalence bias: learning and predicting from imbalanced data
Jul 31, 2021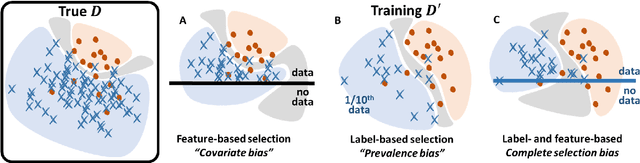
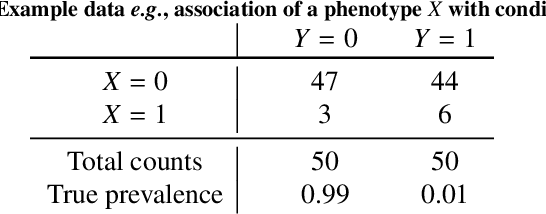

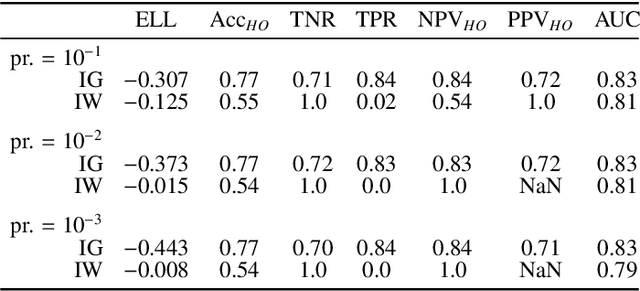
Abstract:Datasets are rarely a realistic approximation of the target population. Say, prevalence is misrepresented, image quality is above clinical standards, etc. This mismatch is known as sampling bias. Sampling biases are a major hindrance for machine learning models. They cause significant gaps between model performance in the lab and in the real world. Our work is a solution to prevalence bias. Prevalence bias is the discrepancy between the prevalence of a pathology and its sampling rate in the training dataset, introduced upon collecting data or due to the practioner rebalancing the training batches. This paper lays the theoretical and computational framework for training models, and for prediction, in the presence of prevalence bias. Concretely a bias-corrected loss function, as well as bias-corrected predictive rules, are derived under the principles of Bayesian risk minimization. The loss exhibits a direct connection to the information gain. It offers a principled alternative to heuristic training losses and complements test-time procedures based on selecting an operating point from summary curves. It integrates seamlessly in the current paradigm of (deep) learning using stochastic backpropagation and naturally with Bayesian models.
A deep learning model for gastric diffuse-type adenocarcinoma classification in whole slide images
Apr 26, 2021



Abstract:Gastric diffuse-type adenocarcinoma represents a disproportionately high percentage of cases of gastric cancers occurring in the young, and its relative incidence seems to be on the rise. Usually it affects the body of the stomach, and presents shorter duration and worse prognosis compared with the differentiated (intestinal) type adenocarcinoma. The main difficulty encountered in the differential diagnosis of gastric adenocarcinomas occurs with the diffuse-type. As the cancer cells of diffuse-type adenocarcinoma are often single and inconspicuous in a background desmoplaia and inflammation, it can often be mistaken for a wide variety of non-neoplastic lesions including gastritis or reactive endothelial cells seen in granulation tissue. In this study we trained deep learning models to classify gastric diffuse-type adenocarcinoma from WSIs. We evaluated the models on five test sets obtained from distinct sources, achieving receiver operator curve (ROC) area under the curves (AUCs) in the range of 0.95-0.99. The highly promising results demonstrate the potential of AI-based computational pathology for aiding pathologists in their diagnostic workflow system.
Partial transfusion: on the expressive influence of trainable batch norm parameters for transfer learning
Feb 10, 2021

Abstract:Transfer learning from ImageNet is the go-to approach when applying deep learning to medical images. The approach is either to fine-tune a pre-trained model or use it as a feature extractor. Most modern architecture contain batch normalisation layers, and fine-tuning a model with such layers requires taking a few precautions as they consist of trainable and non-trainable weights and have two operating modes: training and inference. Attention is primarily given to the non-trainable weights used during inference, as they are the primary source of unexpected behaviour or degradation in performance during transfer learning. It is typically recommended to fine-tune the model with the batch normalisation layers kept in inference mode during both training and inference. In this paper, we pay closer attention instead to the trainable weights of the batch normalisation layers, and we explore their expressive influence in the context of transfer learning. We find that only fine-tuning the trainable weights (scale and centre) of the batch normalisation layers leads to similar performance as to fine-tuning all of the weights, with the added benefit of faster convergence. We demonstrate this on a variety of seven publicly available medical imaging datasets, using four different model architectures.
Deep learning models for gastric signet ring cell carcinoma classification in whole slide images
Nov 18, 2020



Abstract:Signet ring cell carcinoma (SRCC) of the stomach is a rare type of cancer with a slowly rising incidence. It tends to be more difficult to detect by pathologists mainly due to its cellular morphology and diffuse invasion manner, and it has poor prognosis when detected at an advanced stage. Computational pathology tools that can assist pathologists in detecting SRCC would be of a massive benefit. In this paper, we trained deep learning models using transfer learning, fully-supervised learning, and weakly-supervised learning to predict SRCC in Whole Slide Images (WSIs) using a training set of 1,765 WSIs. We evaluated the models on four different test sets of about 500 images each. The best model achieved a Receiver Operator Curve (ROC) area under the curve (AUC) of at least 0.99 on all four test sets, setting a top baseline performance for SRCC WSI classification.
Fully-automated deep learning slice-based muscle estimation from CT images for sarcopenia assessment
Jun 10, 2020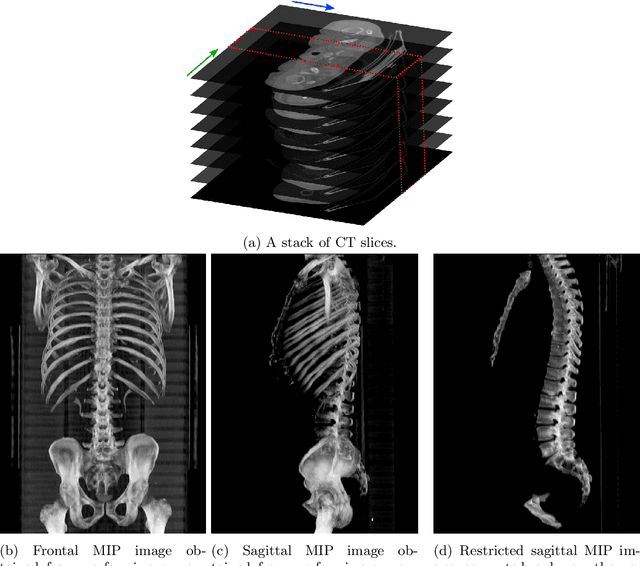
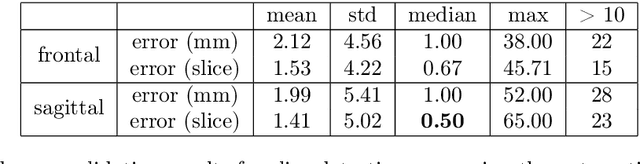
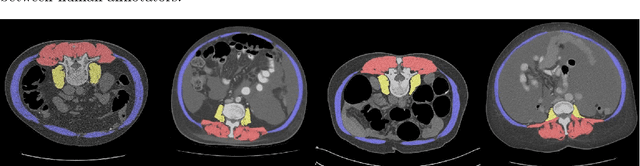

Abstract:Objective: To demonstrate the effectiveness of using a deep learning-based approach for a fully automated slice-based measurement of muscle mass for assessing sarcopenia on CT scans of the abdomen without any case exclusion criteria. Materials and Methods: This retrospective study was conducted using a collection of public and privately available CT images (n = 1070). The method consisted of two stages: slice detection from a CT volume and single-slice CT segmentation. Both stages used Fully Convolutional Neural Networks (FCNN) and were based on a UNet-like architecture. Input data consisted of CT volumes with a variety of fields of view. The output consisted of a segmented muscle mass on a CT slice at the level of L3 vertebra. The muscle mass is segmented into erector spinae, psoas, and rectus abdominus muscle groups. The output was tested against manual ground-truth segmentation by an expert annotator. Results: 3-fold cross validation was used to evaluate the proposed method. The slice detection cross validation error was 1.41+-5.02 (in slices). The segmentation cross validation Dice overlaps were 0.97+-0.02, 0.95+-0.04, 0.94+-0.04 for erector spinae, psoas, and rectus abdominus, respectively, and 0.96+-0.02 for the combined muscle mass. Conclusion: A deep learning approach to detect CT slices and segment muscle mass to perform slice-based analysis of sarcopenia is an effective and promising approach. The use of FCNN to accurately and efficiently detect a slice in CT volumes with a variety of fields of view, occlusions, and slice thicknesses was demonstrated.
Automatic L3 slice detection in 3D CT images using fully-convolutional networks
Nov 22, 2018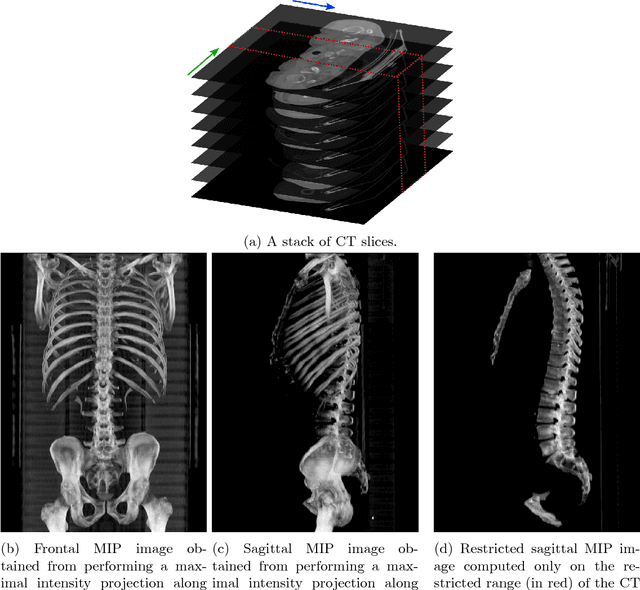


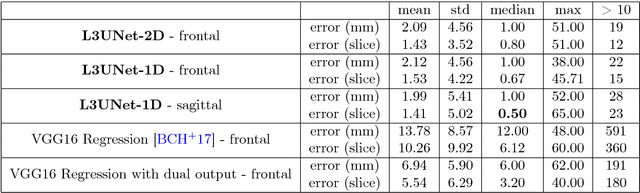
Abstract:The analysis of single CT slices extracted at the third lumbar vertebra (L3) has garnered significant clinical interest in the past few years, in particular in regards to quantifying sarcopenia (muscle loss). In this paper, we propose an efficient method to automatically detect the L3 slice in 3D CT images. Our method works with images with a variety of fields of view, occlusions, and slice thicknesses. 3D CT images are first converted into 2D via Maximal Intensity Projection (MIP), reducing the dimensionality of the problem. The MIP images are then used as input to a 2D fully-convolutional network to predict the L3 slice locations in the form of 2D confidence maps. In addition we propose a variant architecture with less parameters allowing 1D confidence map prediction and slightly faster prediction time without loss of accuracy. Quantitative evaluation of our method on a dataset of 1006 3D CT images yields a median error of 1mm, similar to the inter-rater median error of 1mm obtained from two annotators, demonstrating the effectiveness of our method in efficiently and accurately detecting the L3 slice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge