Shah Islam
Fully-automated deep learning slice-based muscle estimation from CT images for sarcopenia assessment
Jun 10, 2020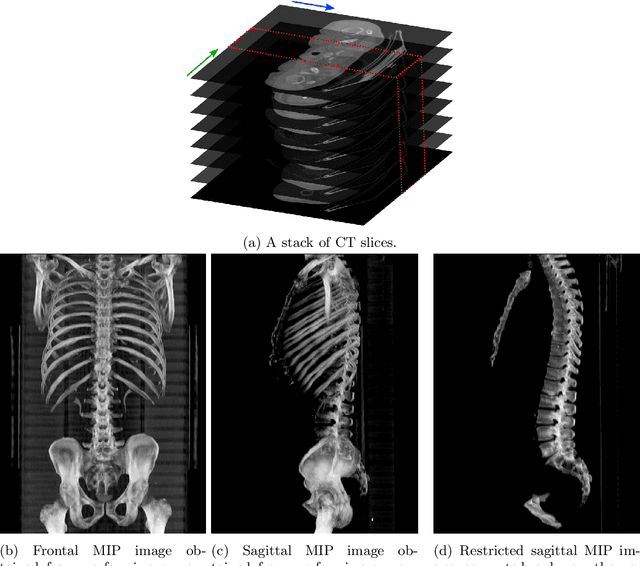
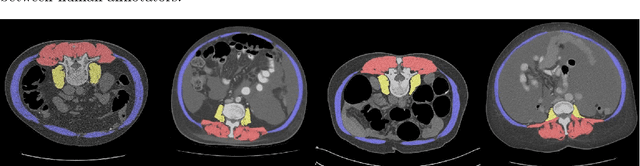

Abstract:Objective: To demonstrate the effectiveness of using a deep learning-based approach for a fully automated slice-based measurement of muscle mass for assessing sarcopenia on CT scans of the abdomen without any case exclusion criteria. Materials and Methods: This retrospective study was conducted using a collection of public and privately available CT images (n = 1070). The method consisted of two stages: slice detection from a CT volume and single-slice CT segmentation. Both stages used Fully Convolutional Neural Networks (FCNN) and were based on a UNet-like architecture. Input data consisted of CT volumes with a variety of fields of view. The output consisted of a segmented muscle mass on a CT slice at the level of L3 vertebra. The muscle mass is segmented into erector spinae, psoas, and rectus abdominus muscle groups. The output was tested against manual ground-truth segmentation by an expert annotator. Results: 3-fold cross validation was used to evaluate the proposed method. The slice detection cross validation error was 1.41+-5.02 (in slices). The segmentation cross validation Dice overlaps were 0.97+-0.02, 0.95+-0.04, 0.94+-0.04 for erector spinae, psoas, and rectus abdominus, respectively, and 0.96+-0.02 for the combined muscle mass. Conclusion: A deep learning approach to detect CT slices and segment muscle mass to perform slice-based analysis of sarcopenia is an effective and promising approach. The use of FCNN to accurately and efficiently detect a slice in CT volumes with a variety of fields of view, occlusions, and slice thicknesses was demonstrated.
Automatic L3 slice detection in 3D CT images using fully-convolutional networks
Nov 22, 2018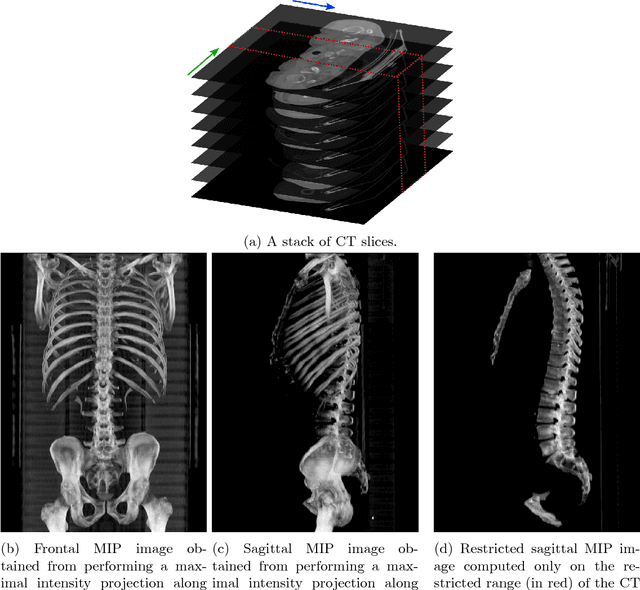


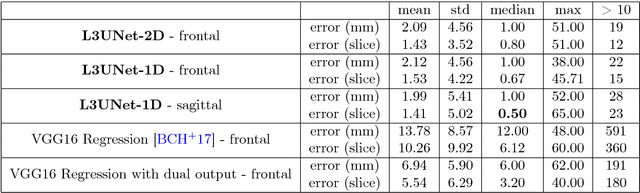
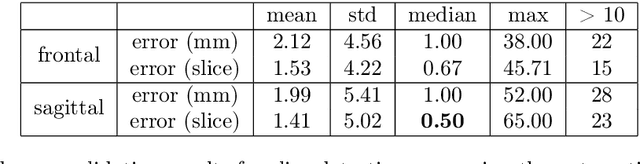
Abstract:The analysis of single CT slices extracted at the third lumbar vertebra (L3) has garnered significant clinical interest in the past few years, in particular in regards to quantifying sarcopenia (muscle loss). In this paper, we propose an efficient method to automatically detect the L3 slice in 3D CT images. Our method works with images with a variety of fields of view, occlusions, and slice thicknesses. 3D CT images are first converted into 2D via Maximal Intensity Projection (MIP), reducing the dimensionality of the problem. The MIP images are then used as input to a 2D fully-convolutional network to predict the L3 slice locations in the form of 2D confidence maps. In addition we propose a variant architecture with less parameters allowing 1D confidence map prediction and slightly faster prediction time without loss of accuracy. Quantitative evaluation of our method on a dataset of 1006 3D CT images yields a median error of 1mm, similar to the inter-rater median error of 1mm obtained from two annotators, demonstrating the effectiveness of our method in efficiently and accurately detecting the L3 slice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge