Eric O. Aboagye
Development of a Multi-Task Learning V-Net for Pulmonary Lobar Segmentation on Computed Tomography and Application to Diseased Lungs
May 11, 2021



Abstract:Automated lobar segmentation allows regional evaluation of lung disease and is important for diagnosis and therapy planning. Advanced statistical workflows permitting such evaluation is a needed area within respiratory medicine; their adoption remains slow, with poor workflow accuracy. Diseased lung regions often produce high-density zones on CT images, limiting an algorithm's execution to specify damaged lobes due to oblique or lacking fissures. This impact motivated developing an improved machine learning method to segment lung lobes that utilises tracheobronchial tree information to enhance segmentation accuracy through the algorithm's spatial familiarity to define lobar extent more accurately. The method undertakes parallel segmentation of lobes and auxiliary tissues simultaneously by employing multi-task learning (MTL) in conjunction with V-Net-attention, a popular convolutional neural network in the imaging realm. In keeping with the model's adeptness for better generalisation, high performance was retained in an external dataset of patients with four distinct diseases: severe lung cancer, COVID-19 pneumonitis, collapsed lungs and Chronic Obstructive Pulmonary Disease (COPD), even though the training data included none of these cases. The benefit of our external validation test is specifically relevant since our choice includes those patients who have diagnosed lung disease with associated radiological abnormalities. To ensure equal rank is given to all segmentations in the main task we report the following performance (Dice score) on a per-segment basis: normal lungs 0.97, COPD 0.94, lung cancer 0.94, COVID-19 pneumonitis 0.94 and collapsed lung 0.92, all at p<0.05. Even segmenting lobes with large deformations on CT images, the model maintained high accuracy. The approach can be readily adopted in the clinical setting as a robust tool for radiologists.
Fully-automated deep learning slice-based muscle estimation from CT images for sarcopenia assessment
Jun 10, 2020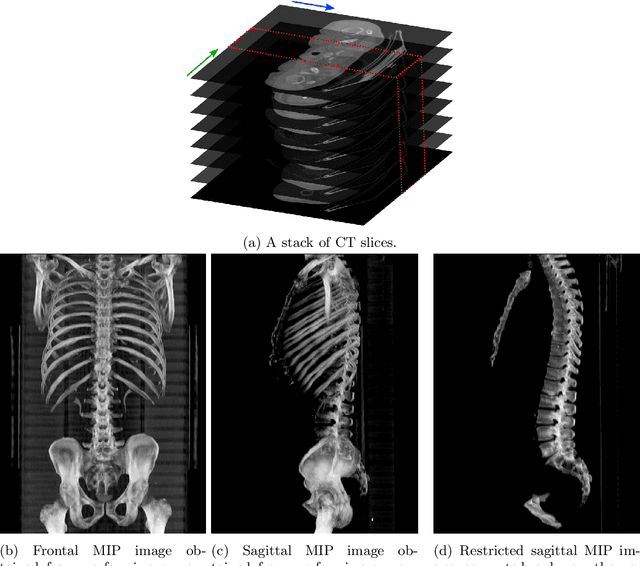
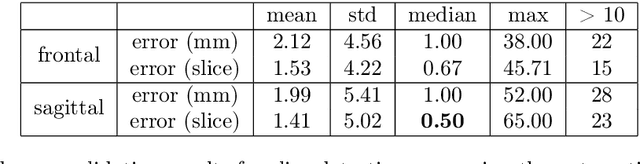
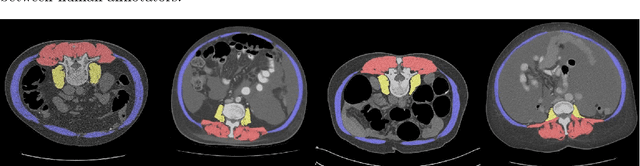

Abstract:Objective: To demonstrate the effectiveness of using a deep learning-based approach for a fully automated slice-based measurement of muscle mass for assessing sarcopenia on CT scans of the abdomen without any case exclusion criteria. Materials and Methods: This retrospective study was conducted using a collection of public and privately available CT images (n = 1070). The method consisted of two stages: slice detection from a CT volume and single-slice CT segmentation. Both stages used Fully Convolutional Neural Networks (FCNN) and were based on a UNet-like architecture. Input data consisted of CT volumes with a variety of fields of view. The output consisted of a segmented muscle mass on a CT slice at the level of L3 vertebra. The muscle mass is segmented into erector spinae, psoas, and rectus abdominus muscle groups. The output was tested against manual ground-truth segmentation by an expert annotator. Results: 3-fold cross validation was used to evaluate the proposed method. The slice detection cross validation error was 1.41+-5.02 (in slices). The segmentation cross validation Dice overlaps were 0.97+-0.02, 0.95+-0.04, 0.94+-0.04 for erector spinae, psoas, and rectus abdominus, respectively, and 0.96+-0.02 for the combined muscle mass. Conclusion: A deep learning approach to detect CT slices and segment muscle mass to perform slice-based analysis of sarcopenia is an effective and promising approach. The use of FCNN to accurately and efficiently detect a slice in CT volumes with a variety of fields of view, occlusions, and slice thicknesses was demonstrated.
Automatic L3 slice detection in 3D CT images using fully-convolutional networks
Nov 22, 2018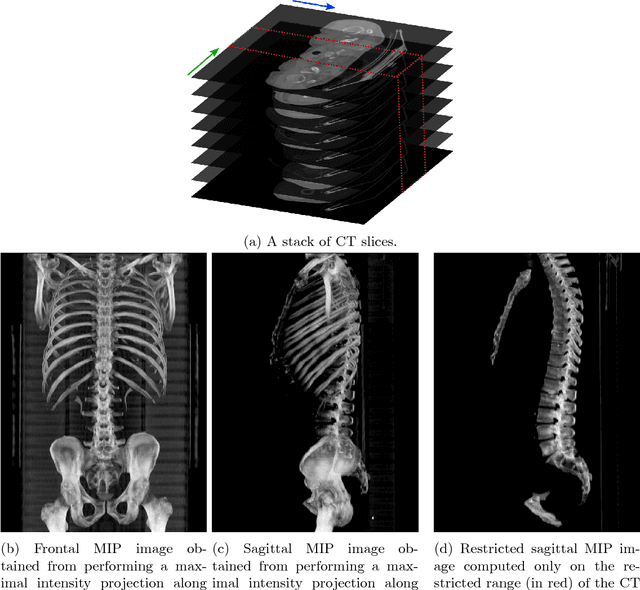


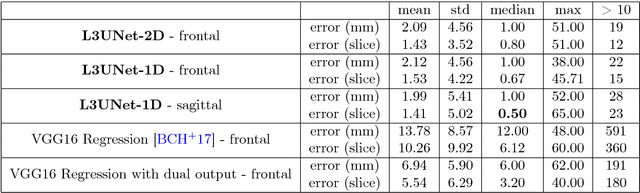
Abstract:The analysis of single CT slices extracted at the third lumbar vertebra (L3) has garnered significant clinical interest in the past few years, in particular in regards to quantifying sarcopenia (muscle loss). In this paper, we propose an efficient method to automatically detect the L3 slice in 3D CT images. Our method works with images with a variety of fields of view, occlusions, and slice thicknesses. 3D CT images are first converted into 2D via Maximal Intensity Projection (MIP), reducing the dimensionality of the problem. The MIP images are then used as input to a 2D fully-convolutional network to predict the L3 slice locations in the form of 2D confidence maps. In addition we propose a variant architecture with less parameters allowing 1D confidence map prediction and slightly faster prediction time without loss of accuracy. Quantitative evaluation of our method on a dataset of 1006 3D CT images yields a median error of 1mm, similar to the inter-rater median error of 1mm obtained from two annotators, demonstrating the effectiveness of our method in efficiently and accurately detecting the L3 slice.
Small Organ Segmentation in Whole-body MRI using a Two-stage FCN and Weighting Schemes
Jul 30, 2018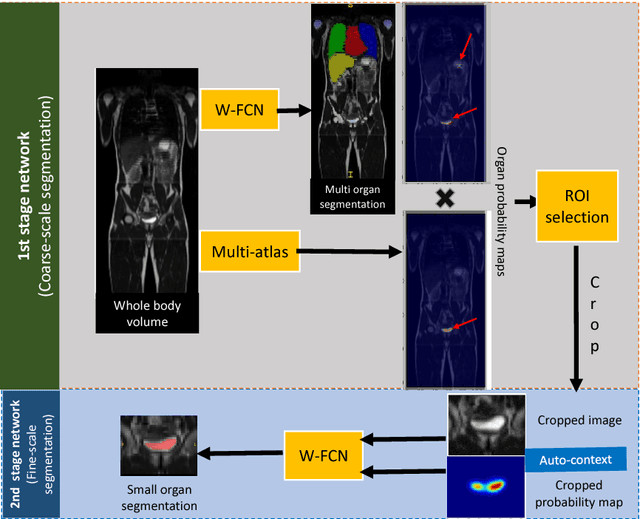

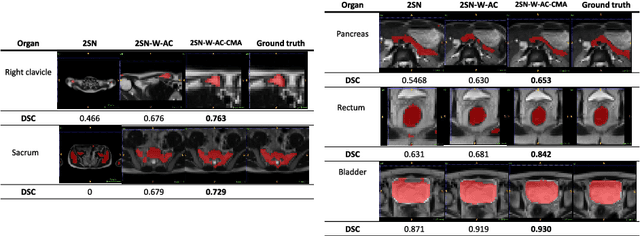
Abstract:Accurate and robust segmentation of small organs in whole-body MRI is difficult due to anatomical variation and class imbalance. Recent deep network based approaches have demonstrated promising performance on abdominal multi-organ segmentations. However, the performance on small organs is still suboptimal as these occupy only small regions of the whole-body volumes with unclear boundaries and variable shapes. A coarse-to-fine, hierarchical strategy is a common approach to alleviate this problem, however, this might miss useful contextual information. We propose a two-stage approach with weighting schemes based on auto-context and spatial atlas priors. Our experiments show that the proposed approach can boost the segmentation accuracy of multiple small organs in whole-body MRI scans.
Domain Adaptation for MRI Organ Segmentation using Reverse Classification Accuracy
Jun 01, 2018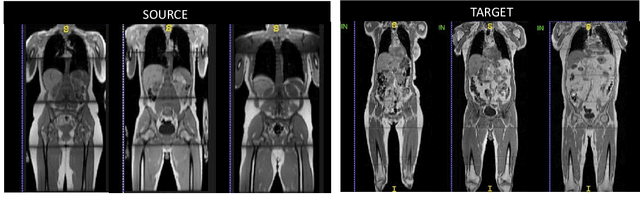

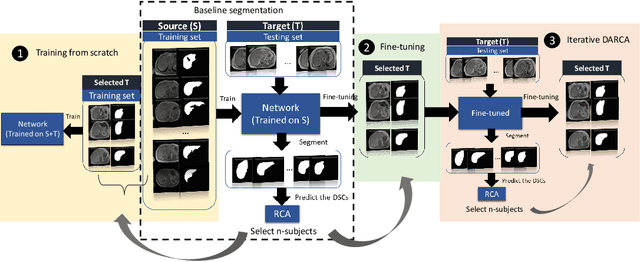

Abstract:The variations in multi-center data in medical imaging studies have brought the necessity of domain adaptation. Despite the advancement of machine learning in automatic segmentation, performance often degrades when algorithms are applied on new data acquired from different scanners or sequences than the training data. Manual annotation is costly and time consuming if it has to be carried out for every new target domain. In this work, we investigate automatic selection of suitable subjects to be annotated for supervised domain adaptation using the concept of reverse classification accuracy (RCA). RCA predicts the performance of a trained model on data from the new domain and different strategies of selecting subjects to be included in the adaptation via transfer learning are evaluated. We perform experiments on a two-center MR database for the task of organ segmentation. We show that subject selection via RCA can reduce the burden of annotation of new data for the target domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge