Tobias Pischon
VERIDAH: Solving Enumeration Anomaly Aware Vertebra Labeling across Imaging Sequences
Jan 20, 2026Abstract:The human spine commonly consists of seven cervical, twelve thoracic, and five lumbar vertebrae. However, enumeration anomalies may result in individuals having eleven or thirteen thoracic vertebrae and four or six lumbar vertebrae. Although the identification of enumeration anomalies has potential clinical implications for chronic back pain and operation planning, the thoracolumbar junction is often poorly assessed and rarely described in clinical reports. Additionally, even though multiple deep-learning-based vertebra labeling algorithms exist, there is a lack of methods to automatically label enumeration anomalies. Our work closes that gap by introducing "Vertebra Identification with Anomaly Handling" (VERIDAH), a novel vertebra labeling algorithm based on multiple classification heads combined with a weighted vertebra sequence prediction algorithm. We show that our approach surpasses existing models on T2w TSE sagittal (98.30% vs. 94.24% of subjects with all vertebrae correctly labeled, p < 0.001) and CT imaging (99.18% vs. 77.26% of subjects with all vertebrae correctly labeled, p < 0.001) and works in arbitrary field-of-view images. VERIDAH correctly labeled the presence 2 Möller et al. of thoracic enumeration anomalies in 87.80% and 96.30% of T2w and CT images, respectively, and lumbar enumeration anomalies in 94.48% and 97.22% for T2w and CT, respectively. Our code and models are available at: https://github.com/Hendrik-code/spineps.
Can synthetic data reproduce real-world findings in epidemiology? A replication study using tree-based generative AI
Aug 19, 2025Abstract:Generative artificial intelligence for synthetic data generation holds substantial potential to address practical challenges in epidemiology. However, many current methods suffer from limited quality, high computational demands, and complexity for non-experts. Furthermore, common evaluation strategies for synthetic data often fail to directly reflect statistical utility. Against this background, a critical underexplored question is whether synthetic data can reliably reproduce key findings from epidemiological research. We propose the use of adversarial random forests (ARF) as an efficient and convenient method for synthesizing tabular epidemiological data. To evaluate its performance, we replicated statistical analyses from six epidemiological publications and compared original with synthetic results. These publications cover blood pressure, anthropometry, myocardial infarction, accelerometry, loneliness, and diabetes, based on data from the German National Cohort (NAKO Gesundheitsstudie), the Bremen STEMI Registry U45 Study, and the Guelph Family Health Study. Additionally, we assessed the impact of dimensionality and variable complexity on synthesis quality by limiting datasets to variables relevant for individual analyses, including necessary derivations. Across all replicated original studies, results from multiple synthetic data replications consistently aligned with original findings. Even for datasets with relatively low sample size-to-dimensionality ratios, the replication outcomes closely matched the original results across various descriptive and inferential analyses. Reducing dimensionality and pre-deriving variables further enhanced both quality and stability of the results.
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025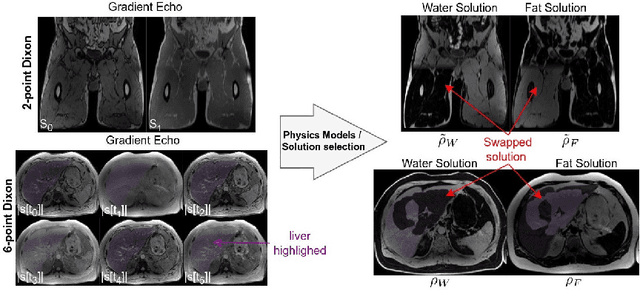
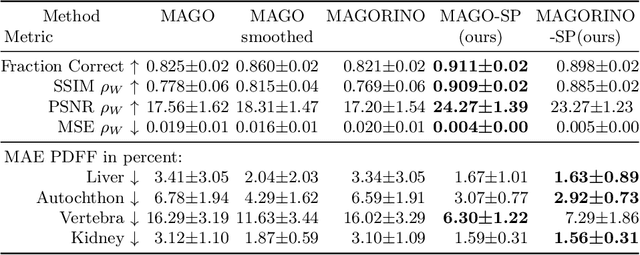
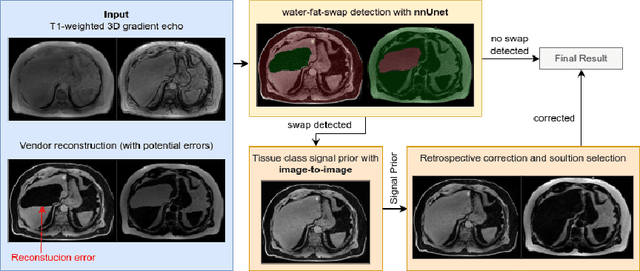
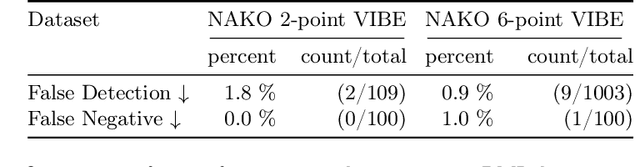
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
Detecting Unforeseen Data Properties with Diffusion Autoencoder Embeddings using Spine MRI data
Oct 14, 2024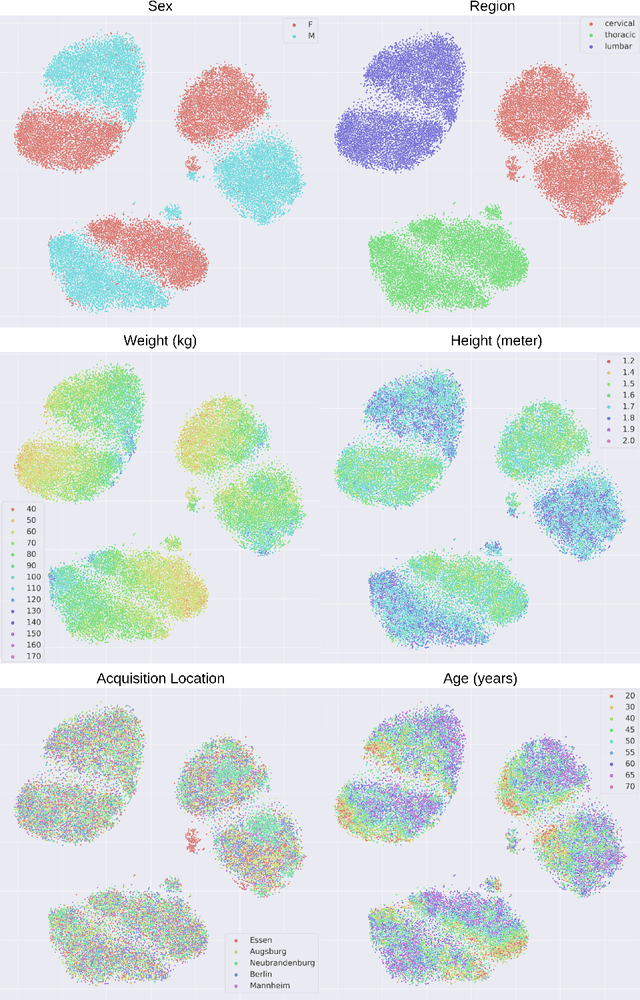

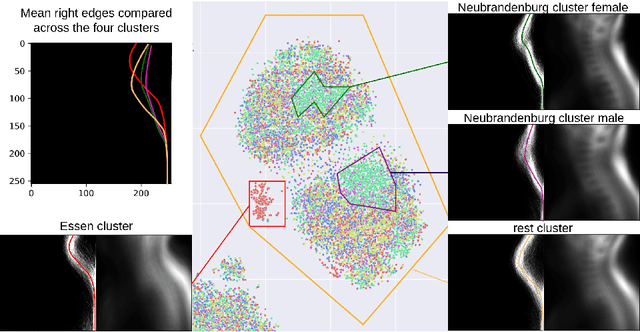
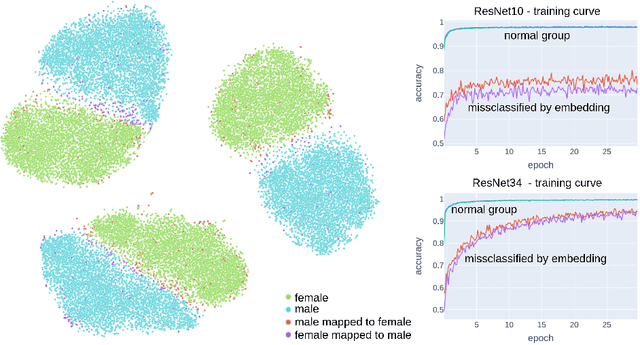
Abstract:Deep learning has made significant strides in medical imaging, leveraging the use of large datasets to improve diagnostics and prognostics. However, large datasets often come with inherent errors through subject selection and acquisition. In this paper, we investigate the use of Diffusion Autoencoder (DAE) embeddings for uncovering and understanding data characteristics and biases, including biases for protected variables like sex and data abnormalities indicative of unwanted protocol variations. We use sagittal T2-weighted magnetic resonance (MR) images of the neck, chest, and lumbar region from 11186 German National Cohort (NAKO) participants. We compare DAE embeddings with existing generative models like StyleGAN and Variational Autoencoder. Evaluations on a large-scale dataset consisting of sagittal T2-weighted MR images of three spine regions show that DAE embeddings effectively separate protected variables such as sex and age. Furthermore, we used t-SNE visualization to identify unwanted variations in imaging protocols, revealing differences in head positioning. Our embedding can identify samples where a sex predictor will have issues learning the correct sex. Our findings highlight the potential of using advanced embedding techniques like DAEs to detect data quality issues and biases in medical imaging datasets. Identifying such hidden relations can enhance the reliability and fairness of deep learning models in healthcare applications, ultimately improving patient care and outcomes.
SPINEPS -- Automatic Whole Spine Segmentation of T2-weighted MR images using a Two-Phase Approach to Multi-class Semantic and Instance Segmentation
Feb 26, 2024
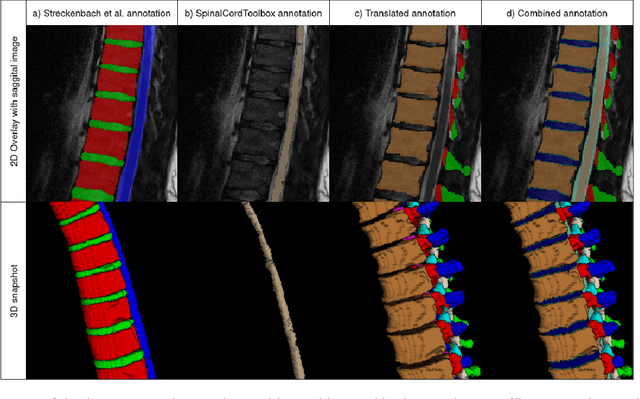
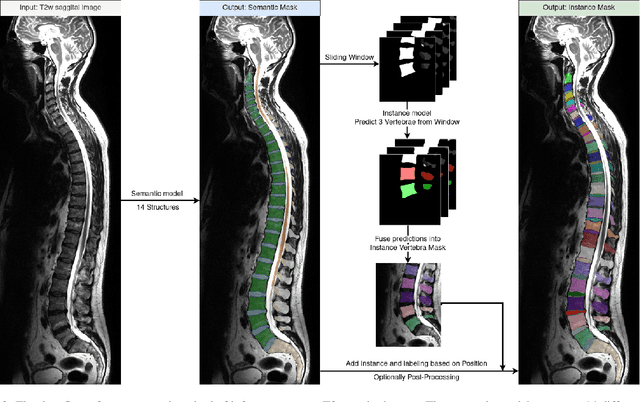
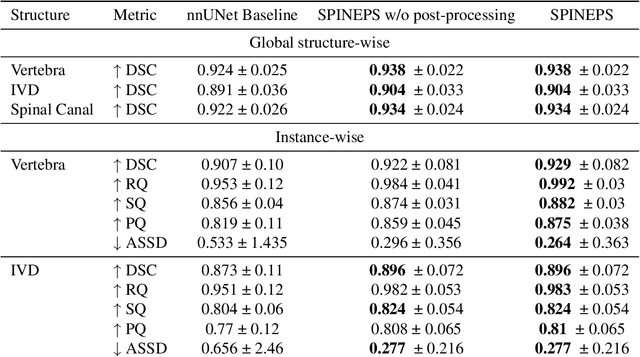
Abstract:Purpose. To present SPINEPS, an open-source deep learning approach for semantic and instance segmentation of 14 spinal structures (ten vertebra substructures, intervertebral discs, spinal cord, spinal canal, and sacrum) in whole body T2w MRI. Methods. During this HIPPA-compliant, retrospective study, we utilized the public SPIDER dataset (218 subjects, 63% female) and a subset of the German National Cohort (1423 subjects, mean age 53, 49% female) for training and evaluation. We combined CT and T2w segmentations to train models that segment 14 spinal structures in T2w sagittal scans both semantically and instance-wise. Performance evaluation metrics included Dice similarity coefficient, average symmetrical surface distance, panoptic quality, segmentation quality, and recognition quality. Statistical significance was assessed using the Wilcoxon signed-rank test. An in-house dataset was used to qualitatively evaluate out-of-distribution samples. Results. On the public dataset, our approach outperformed the baseline (instance-wise vertebra dice score 0.929 vs. 0.907, p-value<0.001). Training on auto-generated annotations and evaluating on manually corrected test data from the GNC yielded global dice scores of 0.900 for vertebrae, 0.960 for intervertebral discs, and 0.947 for the spinal canal. Incorporating the SPIDER dataset during training increased these scores to 0.920, 0.967, 0.958, respectively. Conclusions. The proposed segmentation approach offers robust segmentation of 14 spinal structures in T2w sagittal images, including the spinal cord, spinal canal, intervertebral discs, endplate, sacrum, and vertebrae. The approach yields both a semantic and instance mask as output, thus being easy to utilize. This marks the first publicly available algorithm for whole spine segmentation in sagittal T2w MR imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge