Eimo Martens
Multi-View Stenosis Classification Leveraging Transformer-Based Multiple-Instance Learning Using Real-World Clinical Data
Feb 02, 2026Abstract:Coronary artery stenosis is a leading cause of cardiovascular disease, diagnosed by analyzing the coronary arteries from multiple angiography views. Although numerous deep-learning models have been proposed for stenosis detection from a single angiography view, their performance heavily relies on expensive view-level annotations, which are often not readily available in hospital systems. Moreover, these models fail to capture the temporal dynamics and dependencies among multiple views, which are crucial for clinical diagnosis. To address this, we propose SegmentMIL, a transformer-based multi-view multiple-instance learning framework for patient-level stenosis classification. Trained on a real-world clinical dataset, using patient-level supervision and without any view-level annotations, SegmentMIL jointly predicts the presence of stenosis and localizes the affected anatomical region, distinguishing between the right and left coronary arteries and their respective segments. SegmentMIL obtains high performance on internal and external evaluations and outperforms both view-level models and classical MIL baselines, underscoring its potential as a clinically viable and scalable solution for coronary stenosis diagnosis. Our code is available at https://github.com/NikolaCenic/mil-stenosis.
Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI
Aug 09, 2023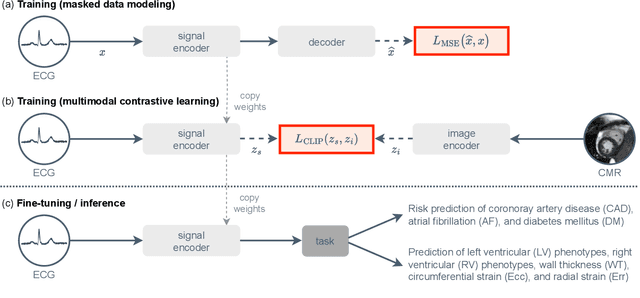
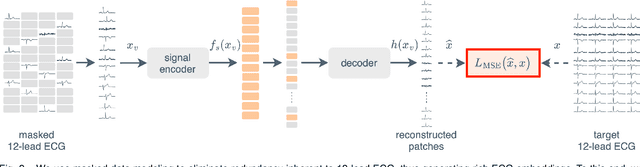
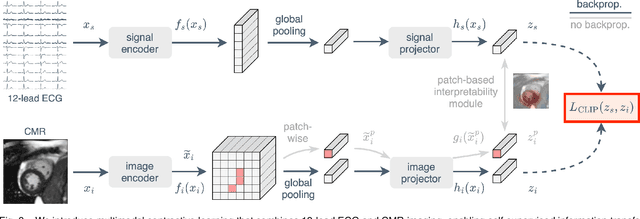
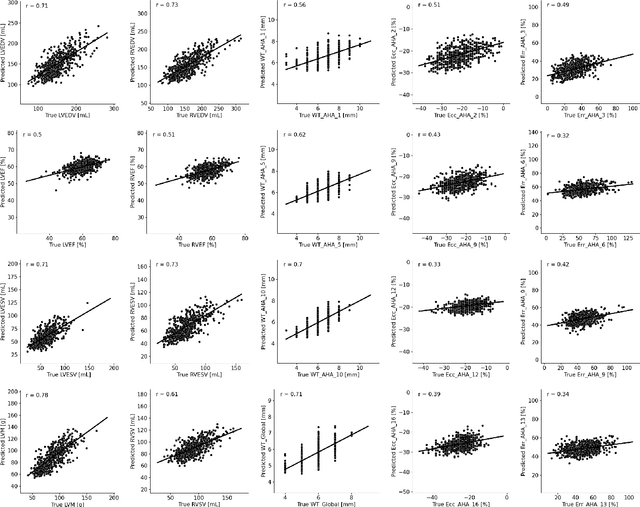
Abstract:The electrocardiogram (ECG) is a widely available diagnostic tool that allows for a cost-effective and fast assessment of the cardiovascular health. However, more detailed examination with expensive cardiac magnetic resonance (CMR) imaging is often preferred for the diagnosis of cardiovascular diseases. While providing detailed visualization of the cardiac anatomy, CMR imaging is not widely available due to long scan times and high costs. To address this issue, we propose the first self-supervised contrastive approach that transfers domain-specific information from CMR images to ECG embeddings. Our approach combines multimodal contrastive learning with masked data modeling to enable holistic cardiac screening solely from ECG data. In extensive experiments using data from 40,044 UK Biobank subjects, we demonstrate the utility and generalizability of our method. We predict the subject-specific risk of various cardiovascular diseases and determine distinct cardiac phenotypes solely from ECG data. In a qualitative analysis, we demonstrate that our learned ECG embeddings incorporate information from CMR image regions of interest. We make our entire pipeline publicly available, including the source code and pre-trained model weights.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge