Peter F. Neher
*: shared first/last authors
Tract orientation mapping for bundle-specific tractography
Jun 14, 2018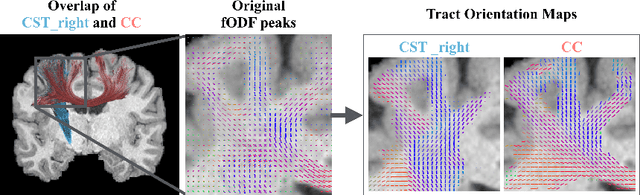
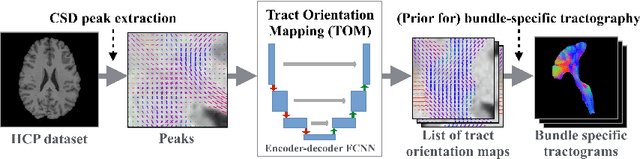
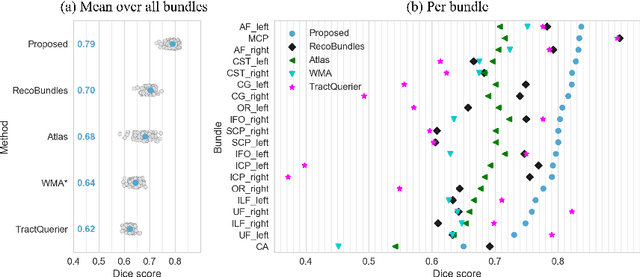
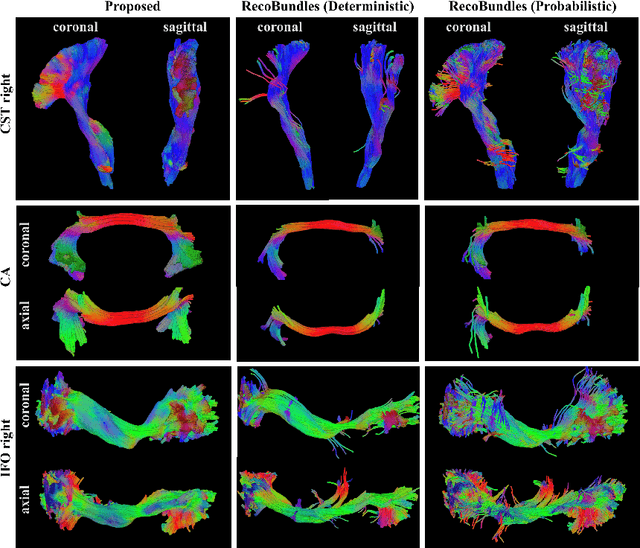
Abstract:While the major white matter tracts are of great interest to numerous studies in neuroscience and medicine, their manual dissection in larger cohorts from diffusion MRI tractograms is time-consuming, requires expert knowledge and is hard to reproduce. Tract orientation mapping (TOM) is a novel concept that facilitates bundle-specific tractography based on a learned mapping from the original fiber orientation distribution function (fODF) peaks to a list of tract orientation maps (also abbr. TOM). Each TOM represents one of the known tracts with each voxel containing no more than one orientation vector. TOMs can act as a prior or even as direct input for tractography. We use an encoder-decoder fully-convolutional neural network architecture to learn the required mapping. In comparison to previous concepts for the reconstruction of specific bundles, the presented one avoids various cumbersome processing steps like whole brain tractography, atlas registration or clustering. We compare it to four state of the art bundle recognition methods on 20 different bundles in a total of 105 subjects from the Human Connectome Project. Results are anatomically convincing even for difficult tracts, while reaching low angular errors, unprecedented runtimes and top accuracy values (Dice). Our code and our data are openly available.
Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions
Jun 06, 2018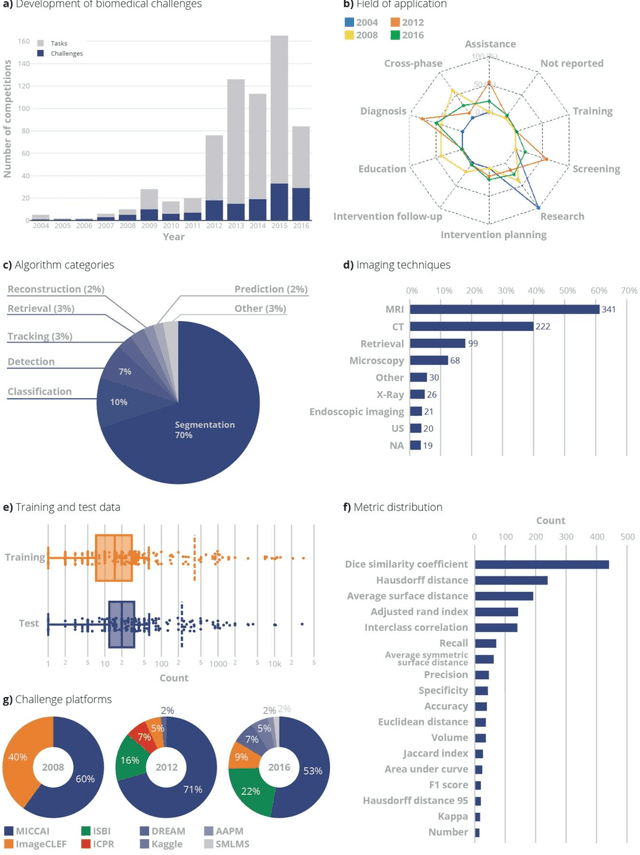
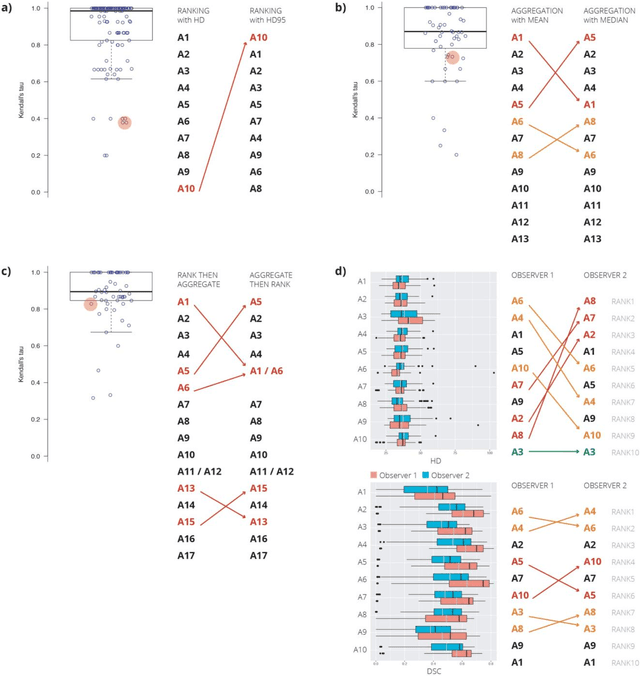
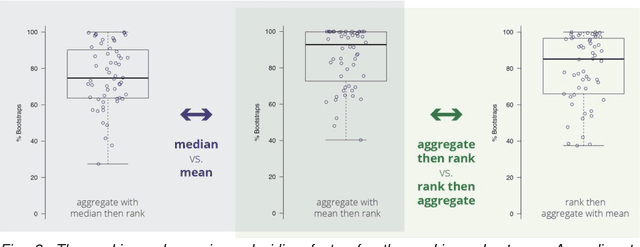
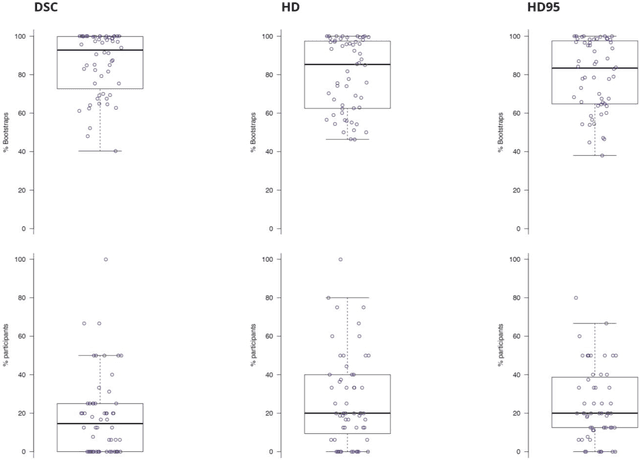
Abstract:International challenges have become the standard for validation of biomedical image analysis methods. Given their scientific impact, it is surprising that a critical analysis of common practices related to the organization of challenges has not yet been performed. In this paper, we present a comprehensive analysis of biomedical image analysis challenges conducted up to now. We demonstrate the importance of challenges and show that the lack of quality control has critical consequences. First, reproducibility and interpretation of the results is often hampered as only a fraction of relevant information is typically provided. Second, the rank of an algorithm is generally not robust to a number of variables such as the test data used for validation, the ranking scheme applied and the observers that make the reference annotations. To overcome these problems, we recommend best practice guidelines and define open research questions to be addressed in the future.
Direct White Matter Bundle Segmentation using Stacked U-Nets
Mar 06, 2017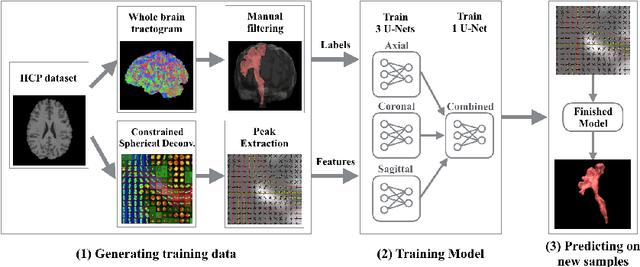
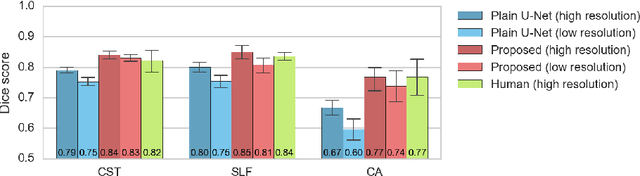
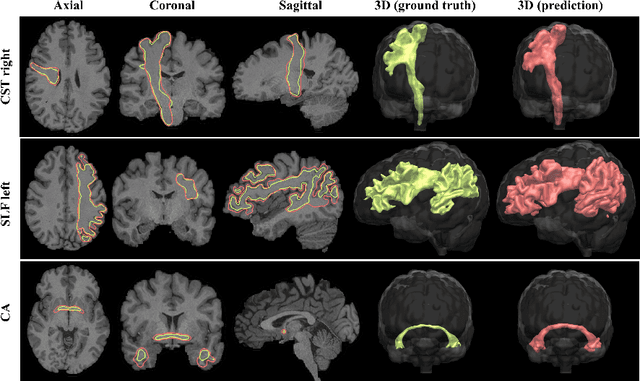
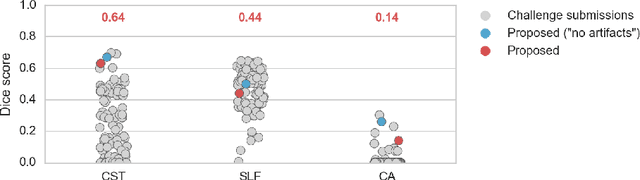
Abstract:The state-of-the-art method for automatically segmenting white matter bundles in diffusion-weighted MRI is tractography in conjunction with streamline cluster selection. This process involves long chains of processing steps which are not only computationally expensive but also complex to setup and tedious with respect to quality control. Direct bundle segmentation methods treat the task as a traditional image segmentation problem. While they so far did not deliver competitive results, they can potentially mitigate many of the mentioned issues. We present a novel supervised approach for direct tract segmentation that shows major performance gains. It builds upon a stacked U-Net architecture which is trained on manual bundle segmentations from Human Connectome Project subjects. We evaluate our approach \textit{in vivo} as well as \textit{in silico} using the ISMRM 2015 Tractography Challenge phantom dataset. We achieve human segmentation performance and a major performance gain over previous pipelines. We show how the learned spatial priors efficiently guide the segmentation even at lower image qualities with little quality loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge