Joshy Cyriac
Multi-centric AI Model for Unruptured Intracranial Aneurysm Detection and Volumetric Segmentation in 3D TOF-MRI
Aug 30, 2024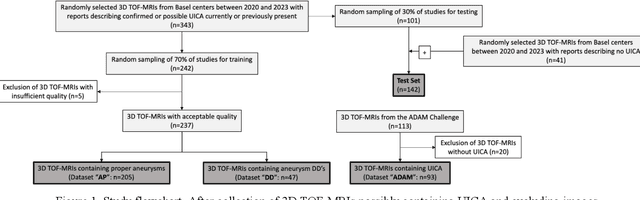
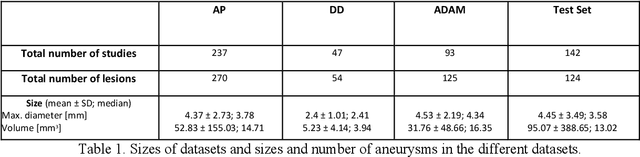
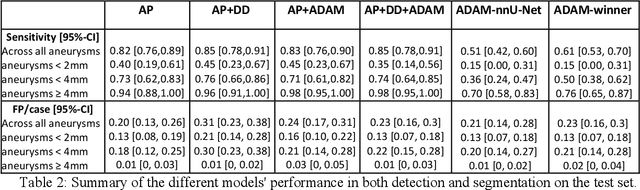
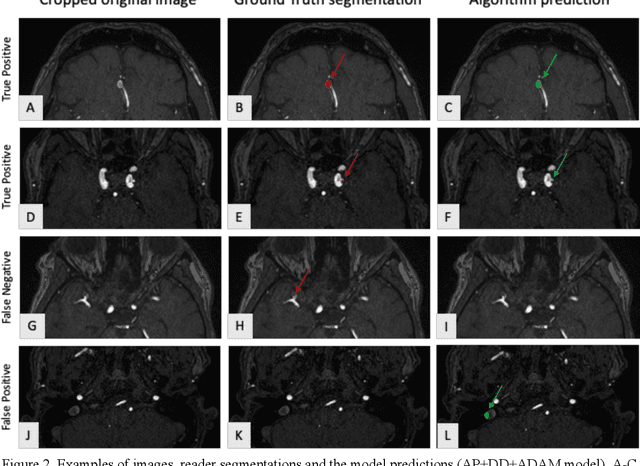
Abstract:Purpose: To develop an open-source nnU-Net-based AI model for combined detection and segmentation of unruptured intracranial aneurysms (UICA) in 3D TOF-MRI, and compare models trained on datasets with aneurysm-like differential diagnoses. Methods: This retrospective study (2020-2023) included 385 anonymized 3D TOF-MRI images from 364 patients (mean age 59 years, 60% female) at multiple centers plus 113 subjects from the ADAM challenge. Images featured untreated or possible UICAs and differential diagnoses. Four distinct training datasets were created, and the nnU-Net framework was used for model development. Performance was assessed on a separate test set using sensitivity and False Positive (FP)/case rate for detection, and DICE score and NSD (Normalized Surface Distance) with a 0.5mm threshold for segmentation. Statistical analysis included chi-square, Mann-Whitney-U, and Kruskal-Wallis tests, with significance set at p < 0.05. Results: Models achieved overall sensitivity between 82% and 85% and a FP/case rate of 0.20 to 0.31, with no significant differences (p = 0.90 and p = 0.16). The primary model showed 85% sensitivity and 0.23 FP/case rate, outperforming the ADAM-challenge winner (61%) and a nnU-Net trained on ADAM data (51%) in sensitivity (p < 0.05). It achieved a mean DICE score of 0.73 and an NSD of 0.84 for correctly detected UICA. Conclusions: Our open-source, nnU-Net-based AI model (available at 10.5281/zenodo.13386859) demonstrates high sensitivity, low false positive rates, and consistent segmentation accuracy for UICA detection and segmentation in 3D TOF-MRI, suggesting its potential to improve clinical diagnosis and for monitoring of UICA.
TotalSegmentator MRI: Sequence-Independent Segmentation of 59 Anatomical Structures in MR images
May 29, 2024



Abstract:Purpose: To develop an open-source and easy-to-use segmentation model that can automatically and robustly segment most major anatomical structures in MR images independently of the MR sequence. Materials and Methods: In this study we extended the capabilities of TotalSegmentator to MR images. 298 MR scans and 227 CT scans were used to segment 59 anatomical structures (20 organs, 18 bones, 11 muscles, 7 vessels, 3 tissue types) relevant for use cases such as organ volumetry, disease characterization, and surgical planning. The MR and CT images were randomly sampled from routine clinical studies and thus represent a real-world dataset (different ages, pathologies, scanners, body parts, sequences, contrasts, echo times, repetition times, field strengths, slice thicknesses and sites). We trained an nnU-Net segmentation algorithm on this dataset and calculated Dice similarity coefficients (Dice) to evaluate the model's performance. Results: The model showed a Dice score of 0.824 (CI: 0.801, 0.842) on the test set, which included a wide range of clinical data with major pathologies. The model significantly outperformed two other publicly available segmentation models (Dice score, 0.824 versus 0.762; p<0.001 and 0.762 versus 0.542; p<0.001). On the CT image test set of the original TotalSegmentator paper it almost matches the performance of the original TotalSegmentator (Dice score, 0.960 versus 0.970; p<0.001). Conclusion: Our proposed model extends the capabilities of TotalSegmentator to MR images. The annotated dataset (https://zenodo.org/doi/10.5281/zenodo.11367004) and open-source toolkit (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
TotalSegmentator: robust segmentation of 104 anatomical structures in CT images
Aug 11, 2022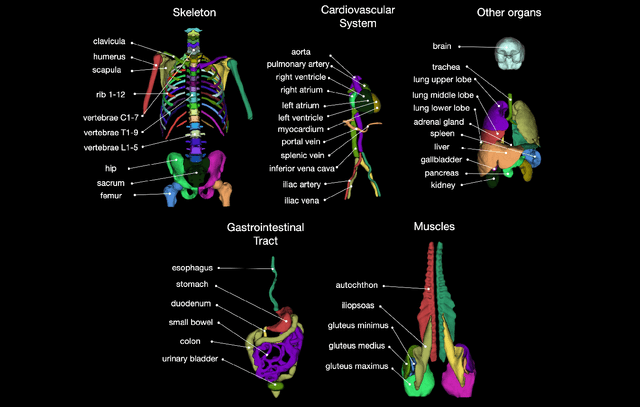
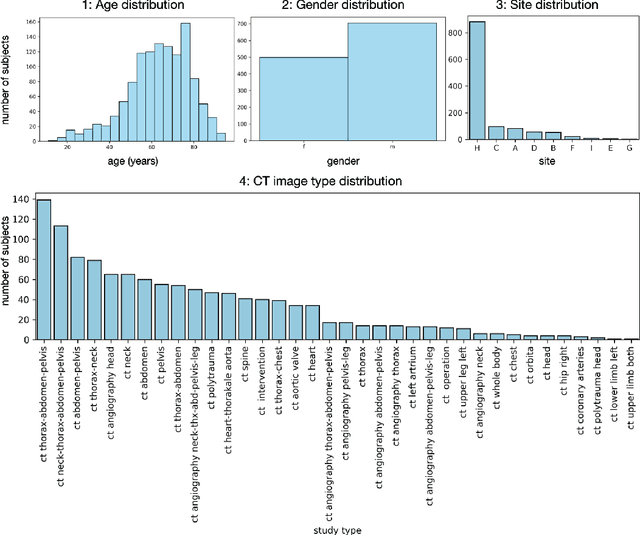
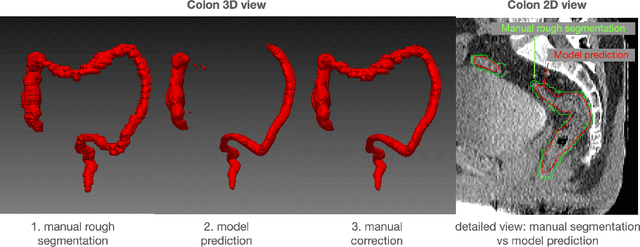
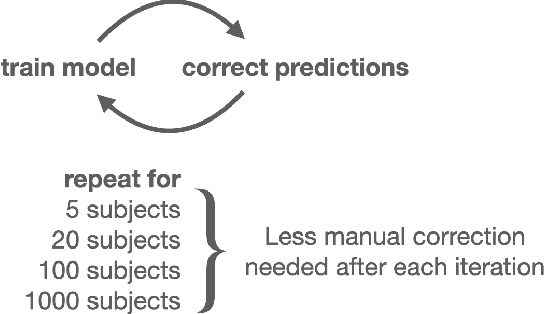
Abstract:In this work we focus on automatic segmentation of multiple anatomical structures in (whole body) CT images. Many segmentation algorithms exist for this task. However, in most cases they suffer from 3 problems: 1. They are difficult to use (the code and data is not publicly available or difficult to use). 2. They do not generalize (often the training dataset was curated to only contain very clean images which do not reflect the image distribution found during clinical routine), 3. The algorithm can only segment one anatomical structure. For more structures several algorithms have to be used which increases the effort required to set up the system. In this work we publish a new dataset and segmentation toolkit which solves all three of these problems: In 1204 CT images we segmented 104 anatomical structures (27 organs, 59 bones, 10 muscles, 8 vessels) covering a majority of relevant classes for most use cases. We show an improved workflow for the creation of ground truth segmentations which speeds up the process by over 10x. The CT images were randomly sampled from clinical routine, thus representing a real world dataset which generalizes to clinical application. The dataset contains a wide range of different pathologies, scanners, sequences and sites. Finally, we train a segmentation algorithm on this new dataset. We call this algorithm TotalSegmentator and make it easily available as a pretrained python pip package (pip install totalsegmentator). Usage is as simple as TotalSegmentator -i ct.nii.gz -o seg and it works well for most CT images. The code is available at https://github.com/wasserth/TotalSegmentator and the dataset at https://doi.org/10.5281/zenodo.6802613.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge