Gabriele Lozupone
Latent Diffusion Autoencoders: Toward Efficient and Meaningful Unsupervised Representation Learning in Medical Imaging
Apr 11, 2025Abstract:This study presents Latent Diffusion Autoencoder (LDAE), a novel encoder-decoder diffusion-based framework for efficient and meaningful unsupervised learning in medical imaging, focusing on Alzheimer disease (AD) using brain MR from the ADNI database as a case study. Unlike conventional diffusion autoencoders operating in image space, LDAE applies the diffusion process in a compressed latent representation, improving computational efficiency and making 3D medical imaging representation learning tractable. To validate the proposed approach, we explore two key hypotheses: (i) LDAE effectively captures meaningful semantic representations on 3D brain MR associated with AD and ageing, and (ii) LDAE achieves high-quality image generation and reconstruction while being computationally efficient. Experimental results support both hypotheses: (i) linear-probe evaluations demonstrate promising diagnostic performance for AD (ROC-AUC: 90%, ACC: 84%) and age prediction (MAE: 4.1 years, RMSE: 5.2 years); (ii) the learned semantic representations enable attribute manipulation, yielding anatomically plausible modifications; (iii) semantic interpolation experiments show strong reconstruction of missing scans, with SSIM of 0.969 (MSE: 0.0019) for a 6-month gap. Even for longer gaps (24 months), the model maintains robust performance (SSIM > 0.93, MSE < 0.004), indicating an ability to capture temporal progression trends; (iv) compared to conventional diffusion autoencoders, LDAE significantly increases inference throughput (20x faster) while also enhancing reconstruction quality. These findings position LDAE as a promising framework for scalable medical imaging applications, with the potential to serve as a foundation model for medical image analysis. Code available at https://github.com/GabrieleLozupone/LDAE
AXIAL: Attention-based eXplainability for Interpretable Alzheimer's Localized Diagnosis using 2D CNNs on 3D MRI brain scans
Jul 02, 2024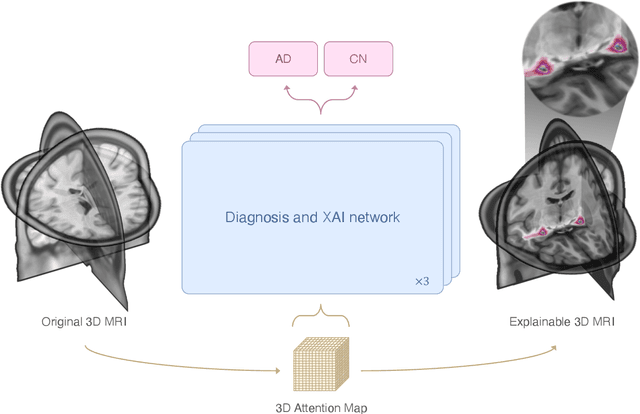
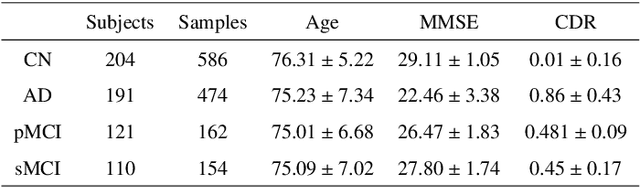
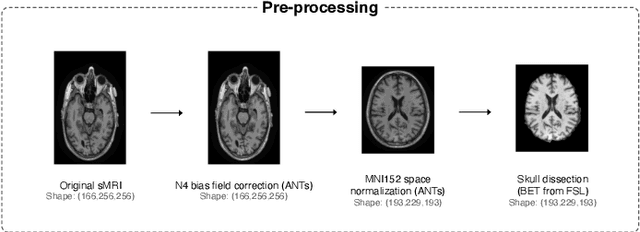
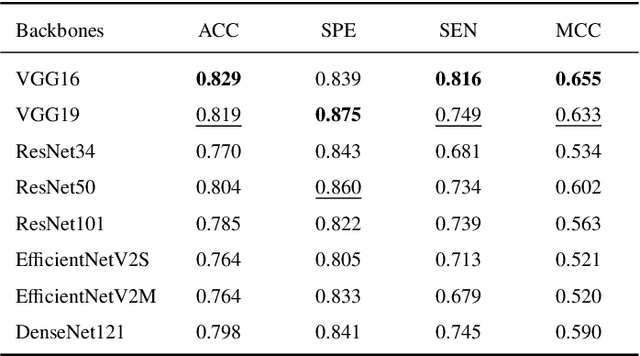
Abstract:This study presents an innovative method for Alzheimer's disease diagnosis using 3D MRI designed to enhance the explainability of model decisions. Our approach adopts a soft attention mechanism, enabling 2D CNNs to extract volumetric representations. At the same time, the importance of each slice in decision-making is learned, allowing the generation of a voxel-level attention map to produces an explainable MRI. To test our method and ensure the reproducibility of our results, we chose a standardized collection of MRI data from the Alzheimer's Disease Neuroimaging Initiative (ADNI). On this dataset, our method significantly outperforms state-of-the-art methods in (i) distinguishing AD from cognitive normal (CN) with an accuracy of 0.856 and Matthew's correlation coefficient (MCC) of 0.712, representing improvements of 2.4\% and 5.3\% respectively over the second-best, and (ii) in the prognostic task of discerning stable from progressive mild cognitive impairment (MCI) with an accuracy of 0.725 and MCC of 0.443, showing improvements of 10.2\% and 20.5\% respectively over the second-best. We achieved this prognostic result by adopting a double transfer learning strategy, which enhanced sensitivity to morphological changes and facilitated early-stage AD detection. With voxel-level precision, our method identified which specific areas are being paid attention to, identifying these predominant brain regions: the \emph{hippocampus}, the \emph{amygdala}, the \emph{parahippocampal}, and the \emph{inferior lateral ventricles}. All these areas are clinically associated with AD development. Furthermore, our approach consistently found the same AD-related areas across different cross-validation folds, proving its robustness and precision in highlighting areas that align closely with known pathological markers of the disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge