Gianluca Pontone
Pericoronary adipose tissue attenuation as a predictor of functional severity of coronary stenosis
Feb 19, 2025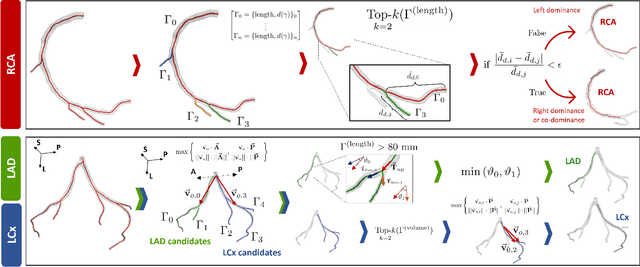
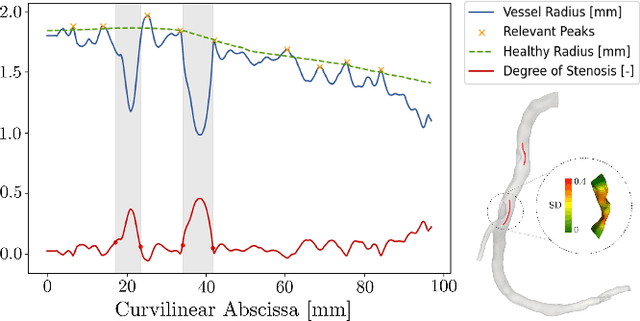
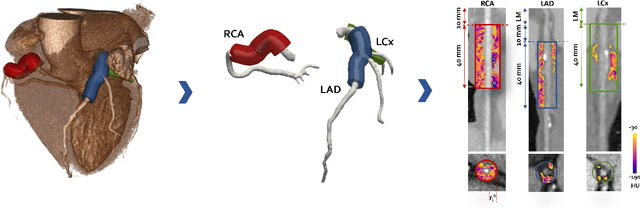
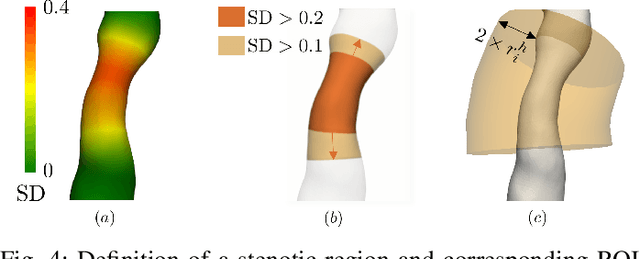
Abstract:Objective: This study aims to evaluate the functional significance of coronary stenosis by analyzing low-level radiomic features of the pericoronary adipose tissue (PCAT) surrounding the lesions, which are indicative of its inflammation status. Methods: A dataset of 72 patients who underwent coronary computed tomography angiography (CCTA) was analyzed, with 3D segmentation and computational fluid dynamics (CFD) simulations from a prior study. Centerlines of the main epicardial branches were automatically extracted, and lesions identified using Gaussian kernel regression to estimate healthy branch caliber. PCAT features were computed per vessel following guideline recommendations and per lesion within a region extending radially for two vessel radii. Features like fat volume and mean attenuation (FAI) were analyzed for their relationship with CFD-derived hemodynamic biomarkers, such as fractional flow reserve (FFR) and wall shear stress (WSS). These features also informed a machine learning (ML) model for classifying potentially ischemic lesions. Results: PCAT exhibited, on average, higher attenuation in the presence of hemodynamically significant lesions (i.e., FFR < 0.80), although this difference was of limited statistical significance. The ML classifier, trained on PCAT features, successfully distinguished potentially ischemic lesions, yielding average accuracy of 0.84. Conclusion: PCAT attenuation is correlated with the functional status of coronary stenosis and can be used to inform ML models for predicting potential ischemia. Significance: PCAT features, readily available from CCTA, can be used to predict the hemodynamic characteristics of a lesion without the need for an invasive FFR examination.
Learning Hemodynamic Scalar Fields on Coronary Artery Meshes: A Benchmark of Geometric Deep Learning Models
Jan 15, 2025



Abstract:Coronary artery disease, caused by the narrowing of coronary vessels due to atherosclerosis, is the leading cause of death worldwide. The diagnostic gold standard, fractional flow reserve (FFR), measures the trans-stenotic pressure ratio during maximal vasodilation but is invasive and costly. This has driven the development of virtual FFR (vFFR) using computational fluid dynamics (CFD) to simulate coronary flow. Geometric deep learning algorithms have shown promise for learning features on meshes, including cardiovascular research applications. This study empirically analyzes various backends for predicting vFFR fields in coronary arteries as CFD surrogates, comparing six backends for learning hemodynamics on meshes using CFD solutions as ground truth. The study has two parts: i) Using 1,500 synthetic left coronary artery bifurcations, models were trained to predict pressure-related fields for vFFR reconstruction, comparing different learning variables. ii) Using 427 patient-specific CFD simulations, experiments were repeated focusing on the best-performing learning variable from the synthetic dataset. Most backends performed well on the synthetic dataset, especially when predicting pressure drop over the manifold. Transformer-based backends outperformed others when predicting pressure and vFFR fields and were the only models achieving strong performance on patient-specific data, excelling in both average per-point error and vFFR accuracy in stenotic lesions. These results suggest geometric deep learning backends can effectively replace CFD for simple geometries, while transformer-based networks are superior for complex, heterogeneous datasets. Pressure drop was identified as the optimal network output for learning pressure-related fields.
Deep vectorised operators for pulsatile hemodynamics estimation in coronary arteries from a steady-state prior
Oct 15, 2024



Abstract:Cardiovascular hemodynamic fields provide valuable medical decision markers for coronary artery disease. Computational fluid dynamics (CFD) is the gold standard for accurate, non-invasive evaluation of these quantities in vivo. In this work, we propose a time-efficient surrogate model, powered by machine learning, for the estimation of pulsatile hemodynamics based on steady-state priors. We introduce deep vectorised operators, a modelling framework for discretisation independent learning on infinite-dimensional function spaces. The underlying neural architecture is a neural field conditioned on hemodynamic boundary conditions. Importantly, we show how relaxing the requirement of point-wise action to permutation-equivariance leads to a family of models that can be parametrised by message passing and self-attention layers. We evaluate our approach on a dataset of 74 stenotic coronary arteries extracted from coronary computed tomography angiography (CCTA) with patient-specific pulsatile CFD simulations as ground truth. We show that our model produces accurate estimates of the pulsatile velocity and pressure while being agnostic to re-sampling of the source domain (discretisation independence). This shows that deep vectorised operators are a powerful modelling tool for cardiovascular hemodynamics estimation in coronary arteries and beyond.
CAD-RADS scoring of coronary CT angiography with Multi-Axis Vision Transformer: a clinically-inspired deep learning pipeline
Apr 14, 2023Abstract:The standard non-invasive imaging technique used to assess the severity and extent of Coronary Artery Disease (CAD) is Coronary Computed Tomography Angiography (CCTA). However, manual grading of each patient's CCTA according to the CAD-Reporting and Data System (CAD-RADS) scoring is time-consuming and operator-dependent, especially in borderline cases. This work proposes a fully automated, and visually explainable, deep learning pipeline to be used as a decision support system for the CAD screening procedure. The pipeline performs two classification tasks: firstly, identifying patients who require further clinical investigations and secondly, classifying patients into subgroups based on the degree of stenosis, according to commonly used CAD-RADS thresholds. The pipeline pre-processes multiplanar projections of the coronary arteries, extracted from the original CCTAs, and classifies them using a fine-tuned Multi-Axis Vision Transformer architecture. With the aim of emulating the current clinical practice, the model is trained to assign a per-patient score by stacking the bi-dimensional longitudinal cross-sections of the three main coronary arteries along channel dimension. Furthermore, it generates visually interpretable maps to assess the reliability of the predictions. When run on a database of 1873 three-channel images of 253 patients collected at the Monzino Cardiology Center in Milan, the pipeline obtained an AUC of 0.87 and 0.93 for the two classification tasks, respectively. According to our knowledge, this is the first model trained to assign CAD-RADS scores learning solely from patient scores and not requiring finer imaging annotation steps that are not part of the clinical routine.
Rapid quantification of COVID-19 pneumonia burden from computed tomography with convolutional LSTM networks
Mar 31, 2021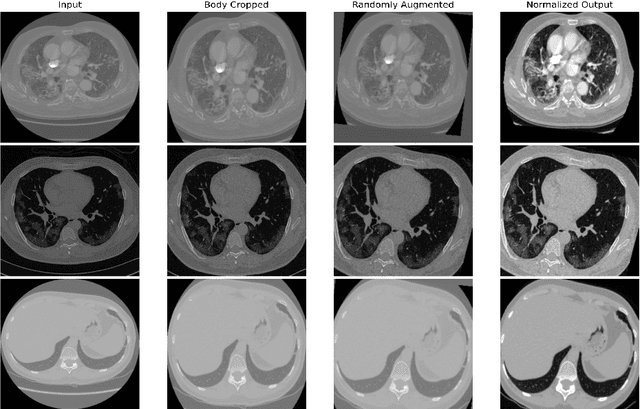
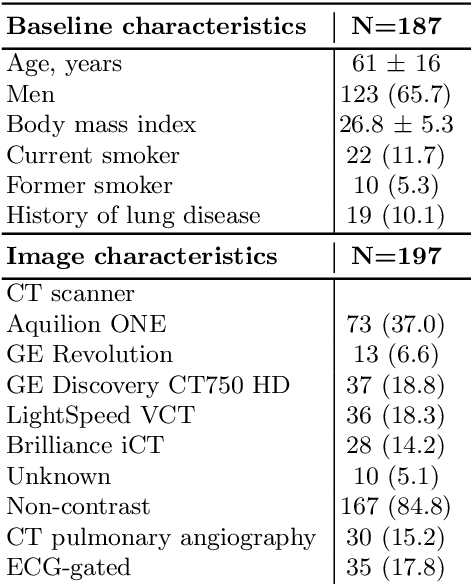
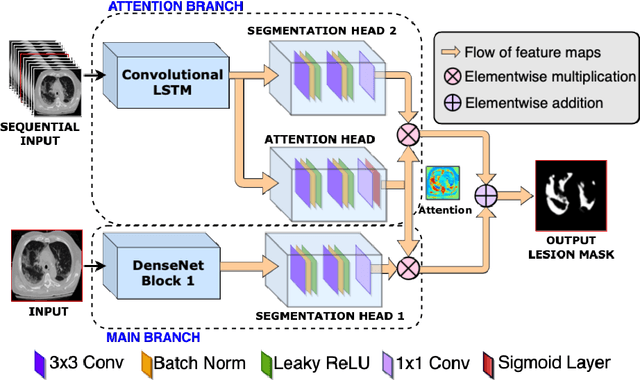
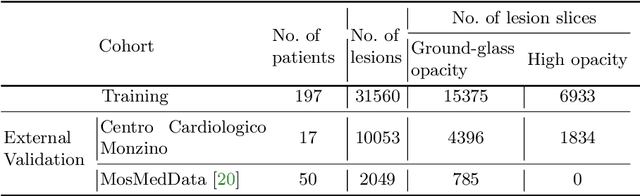
Abstract:Quantitative lung measures derived from computed tomography (CT) have been demonstrated to improve prognostication in coronavirus disease (COVID-19) patients, but are not part of the clinical routine since required manual segmentation of lung lesions is prohibitively time-consuming. We propose a new fully automated deep learning framework for rapid quantification and differentiation between lung lesions in COVID-19 pneumonia from both contrast and non-contrast CT images using convolutional Long Short-Term Memory (ConvLSTM) networks. Utilizing the expert annotations, model training was performed 5 times with separate hold-out sets using 5-fold cross-validation to segment ground-glass opacity and high opacity (including consolidation and pleural effusion). The performance of the method was evaluated on CT data sets from 197 patients with positive reverse transcription polymerase chain reaction test result for SARS-CoV-2. Strong agreement between expert manual and automatic segmentation was obtained for lung lesions with a Dice score coefficient of 0.876 $\pm$ 0.005; excellent correlations of 0.978 and 0.981 for ground-glass opacity and high opacity volumes. In the external validation set of 67 patients, there was dice score coefficient of 0.767 $\pm$ 0.009 as well as excellent correlations of 0.989 and 0.996 for ground-glass opacity and high opacity volumes. Computations for a CT scan comprising 120 slices were performed under 2 seconds on a personal computer equipped with NVIDIA Titan RTX graphics processing unit. Therefore, our deep learning-based method allows rapid fully-automated quantitative measurement of pneumonia burden from CT and may generate results with an accuracy similar to the expert readers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge