Simone Saitta
Attenuation artifact detection and severity classification in intracoronary OCT using mixed image representations
Mar 07, 2025Abstract:In intracoronary optical coherence tomography (OCT), blood residues and gas bubbles cause attenuation artifacts that can obscure critical vessel structures. The presence and severity of these artifacts may warrant re-acquisition, prolonging procedure time and increasing use of contrast agent. Accurate detection of these artifacts can guide targeted re-acquisition, reducing the amount of repeated scans needed to achieve diagnostically viable images. However, the highly heterogeneous appearance of these artifacts poses a challenge for the automated detection of the affected image regions. To enable automatic detection of the attenuation artifacts caused by blood residues and gas bubbles based on their severity, we propose a convolutional neural network that performs classification of the attenuation lines (A-lines) into three classes: no artifact, mild artifact and severe artifact. Our model extracts and merges features from OCT images in both Cartesian and polar coordinates, where each column of the image represents an A-line. Our method detects the presence of attenuation artifacts in OCT frames reaching F-scores of 0.77 and 0.94 for mild and severe artifacts, respectively. The inference time over a full OCT scan is approximately 6 seconds. Our experiments show that analysis of images represented in both Cartesian and polar coordinate systems outperforms the analysis in polar coordinates only, suggesting that these representations contain complementary features. This work lays the foundation for automated artifact assessment and image acquisition guidance in intracoronary OCT imaging.
Pericoronary adipose tissue attenuation as a predictor of functional severity of coronary stenosis
Feb 19, 2025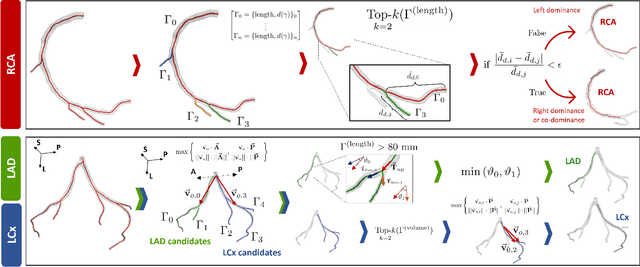
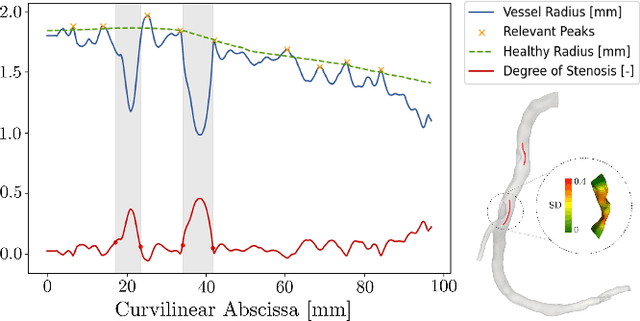
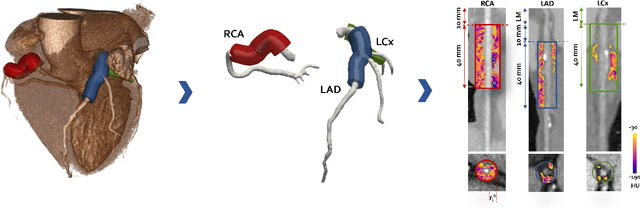
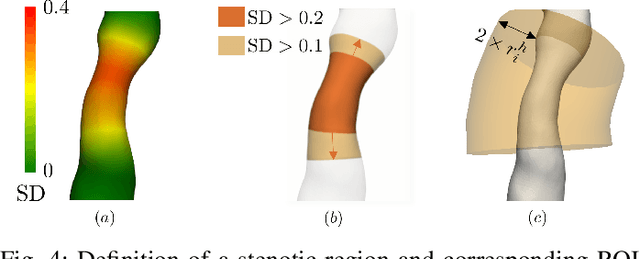
Abstract:Objective: This study aims to evaluate the functional significance of coronary stenosis by analyzing low-level radiomic features of the pericoronary adipose tissue (PCAT) surrounding the lesions, which are indicative of its inflammation status. Methods: A dataset of 72 patients who underwent coronary computed tomography angiography (CCTA) was analyzed, with 3D segmentation and computational fluid dynamics (CFD) simulations from a prior study. Centerlines of the main epicardial branches were automatically extracted, and lesions identified using Gaussian kernel regression to estimate healthy branch caliber. PCAT features were computed per vessel following guideline recommendations and per lesion within a region extending radially for two vessel radii. Features like fat volume and mean attenuation (FAI) were analyzed for their relationship with CFD-derived hemodynamic biomarkers, such as fractional flow reserve (FFR) and wall shear stress (WSS). These features also informed a machine learning (ML) model for classifying potentially ischemic lesions. Results: PCAT exhibited, on average, higher attenuation in the presence of hemodynamically significant lesions (i.e., FFR < 0.80), although this difference was of limited statistical significance. The ML classifier, trained on PCAT features, successfully distinguished potentially ischemic lesions, yielding average accuracy of 0.84. Conclusion: PCAT attenuation is correlated with the functional status of coronary stenosis and can be used to inform ML models for predicting potential ischemia. Significance: PCAT features, readily available from CCTA, can be used to predict the hemodynamic characteristics of a lesion without the need for an invasive FFR examination.
Learning Hemodynamic Scalar Fields on Coronary Artery Meshes: A Benchmark of Geometric Deep Learning Models
Jan 15, 2025



Abstract:Coronary artery disease, caused by the narrowing of coronary vessels due to atherosclerosis, is the leading cause of death worldwide. The diagnostic gold standard, fractional flow reserve (FFR), measures the trans-stenotic pressure ratio during maximal vasodilation but is invasive and costly. This has driven the development of virtual FFR (vFFR) using computational fluid dynamics (CFD) to simulate coronary flow. Geometric deep learning algorithms have shown promise for learning features on meshes, including cardiovascular research applications. This study empirically analyzes various backends for predicting vFFR fields in coronary arteries as CFD surrogates, comparing six backends for learning hemodynamics on meshes using CFD solutions as ground truth. The study has two parts: i) Using 1,500 synthetic left coronary artery bifurcations, models were trained to predict pressure-related fields for vFFR reconstruction, comparing different learning variables. ii) Using 427 patient-specific CFD simulations, experiments were repeated focusing on the best-performing learning variable from the synthetic dataset. Most backends performed well on the synthetic dataset, especially when predicting pressure drop over the manifold. Transformer-based backends outperformed others when predicting pressure and vFFR fields and were the only models achieving strong performance on patient-specific data, excelling in both average per-point error and vFFR accuracy in stenotic lesions. These results suggest geometric deep learning backends can effectively replace CFD for simple geometries, while transformer-based networks are superior for complex, heterogeneous datasets. Pressure drop was identified as the optimal network output for learning pressure-related fields.
World of Forms: Deformable Geometric Templates for One-Shot Surface Meshing in Coronary CT Angiography
Sep 18, 2024Abstract:Deep learning-based medical image segmentation and surface mesh generation typically involve a sequential pipeline from image to segmentation to meshes, often requiring large training datasets while making limited use of prior geometric knowledge. This may lead to topological inconsistencies and suboptimal performance in low-data regimes. To address these challenges, we propose a data-efficient deep learning method for direct 3D anatomical object surface meshing using geometric priors. Our approach employs a multi-resolution graph neural network that operates on a prior geometric template which is deformed to fit object boundaries of interest. We show how different templates may be used for the different surface meshing targets, and introduce a novel masked autoencoder pretraining strategy for 3D spherical data. The proposed method outperforms nnUNet in a one-shot setting for segmentation of the pericardium, left ventricle (LV) cavity and the LV myocardium. Similarly, the method outperforms other lumen segmentation operating on multi-planar reformatted images. Results further indicate that mesh quality is on par with or improves upon marching cubes post-processing of voxel mask predictions, while remaining flexible in the choice of mesh triangulation prior, thus paving the way for more accurate and topologically consistent 3D medical object surface meshing.
Implicit neural representations for unsupervised super-resolution and denoising of 4D flow MRI
Feb 24, 2023



Abstract:4D flow MRI is a non-invasive imaging method that can measure blood flow velocities over time. However, the velocity fields detected by this technique have limitations due to low resolution and measurement noise. Coordinate-based neural networks have been researched to improve accuracy, with SIRENs being suitable for super-resolution tasks. Our study investigates SIRENs for time-varying 3-directional velocity fields measured in the aorta by 4D flow MRI, achieving denoising and super-resolution. We trained our method on voxel coordinates and benchmarked our approach using synthetic measurements and a real 4D flow MRI scan. Our optimized SIREN architecture outperformed state-of-the-art techniques, producing denoised and super-resolved velocity fields from clinical data. Our approach is quick to execute and straightforward to implement for novel cases, achieving 4D super-resolution.
A Deep Learning-Based and Fully Automated Pipeline for Regurgitant Mitral Valve Anatomy Analysis from 3D Echocardiography
Feb 21, 2023Abstract:3D transesophageal echocardiography (3DTEE), is the recommended method for diagnosing mitral regurgitation (MR). 3DTEE provides a high-quality 3D image of the mitral valve (MV), allowing for precise segmentation and measurement of the regurgitant valve anatomy. However, manual TEE segmentations are time-consuming and prone to intra-operator variability, affecting the reliability of the measurements. To address this, we developed a fully automated pipeline using a 3D convolutional neural network (CNN) to segment MV substructures (annulus, anterior leaflet, and posterior leaflet) and quantify MV anatomy. The 3D CNN, based on a multi-decoder residual U-Net architecture, was trained and tested on a dataset comprising 100 3DTEE images with corresponding segmentations. Within the pipeline, a custom algorithm refines the CNN-based segmentations and extracts MV models, from which anatomical landmarks and features are quantified. The accuracy of the proposed method was assessed using Dice score and mean surface distance (MSD) against ground truth segmentations, and the extracted anatomical parameters were compared against a semiautomated commercial software TomTec Image Arena. The trained 3D CNN achieved an average Dice score of 0.79 and MSD of 0.47 mm for the combined segmentation of the annulus, anterior and posterior leaflet. The proposed CNN architecture outperformed a baseline residual U-Net architecture in MV substructure segmentation, and the refinement of the predicted annulus segmentation improved MSD by 8.36%. The annular and leaflet linear measurements differed by less than 7.94 mm and 3.67 mm, respectively, compared to the 3D measurements obtained with TomTec Image Arena. The proposed pipeline was faster than the commercial software, with a modeling time of 12.54 s and a quantification time of 54.42 s.
A CT-based deep learning system for automatic assessment of aortic root morphology for TAVI planning
Feb 10, 2023



Abstract:Accurate planning of transcatheter aortic implantation (TAVI) is important to minimize complications, and it requires anatomic evaluation of the aortic root (AR), commonly done through 3D computed tomography (CT) image analysis. Currently, there is no standard automated solution for this process. Two convolutional neural networks (CNNs) with 3D U-Net architectures (model 1 and model 2) were trained on 310 CT scans for AR analysis. Model 1 performed AR segmentation and model 2 identified the aortic annulus and sinotubular junction (STJ) contours. Results were validated against manual measurements of 178 TAVI candidates. After training, the two models were integrated into a fully automated pipeline for geometric analysis of the AR. The trained CNNs effectively segmented the AR, annulus and STJ, resulting in mean Dice scores of 0.93 for the AR, and mean surface distances of 1.16 mm and 1.30 mm for the annulus and STJ, respectively. Automatic measurements were in good agreement with manual annotations, yielding annulus diameters that differed by 0.52 [-2.96, 4.00] mm (bias and 95% limits of agreement for manual minus algorithm). Evaluating the area-derived diameter, bias and limits of agreement were 0.07 [-0.25, 0.39] mm. STJ and sinuses diameters computed by the automatic method yielded differences of 0.16 [-2.03, 2.34] and 0.1 [-2.93, 3.13] mm, respectively. The proposed tool is a fully automatic solution to quantify morphological biomarkers for pre-TAVI planning. The method was validated against manual annotation from clinical experts and showed to be quick and effective in assessing AR anatomy, with potential for time and cost savings.
Data-driven generation of 4D velocity profiles in the aneurysmal ascending aorta
Nov 01, 2022Abstract:Numerical simulations of blood flow are a valuable tool to investigate the pathophysiology of ascending thoracic aortic aneurysms (ATAA). To accurately reproduce hemodynamics, computational fluid dynamics (CFD) models must employ realistic inflow boundary conditions (BCs). However, the limited availability of in vivo velocity measurements still makes researchers resort to idealized BCs. In this study we generated and thoroughly characterized a large dataset of synthetic 4D aortic velocity profiles suitable to be used as BCs for CFD simulations. 4D flow MRI scans of 30 subjects with ATAA were processed to extract cross-sectional planes along the ascending aorta, ensuring spatial alignment among all planes and interpolating all velocity fields to a reference configuration. Velocity profiles of the clinical cohort were extensively characterized by computing flow morphology descriptors of both spatial and temporal features. By exploiting principal component analysis (PCA), a statistical shape model (SSM) of 4D aortic velocity profiles was built and a dataset of 437 synthetic cases with realistic properties was generated. Comparison between clinical and synthetic datasets showed that the synthetic data presented similar characteristics as the clinical population in terms of key morphological parameters. The average velocity profile qualitatively resembled a parabolic-shaped profile, but was quantitatively characterized by more complex flow patterns which an idealized profile would not replicate. Statistically significant correlations were found between PCA principal modes of variation and flow descriptors. We built a data-driven generative model of 4D aortic velocity profiles, suitable to be used in computational studies of blood flow. The proposed software system also allows to map any of the generated velocity profiles to the inlet plane of any virtual subject given its coordinate set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge