Jonathan W. Weinsaft
Improved Robustness for Deep Learning-based Segmentation of Multi-Center Myocardial Perfusion MRI Datasets Using Data Adaptive Uncertainty-guided Space-time Analysis
Aug 09, 2024


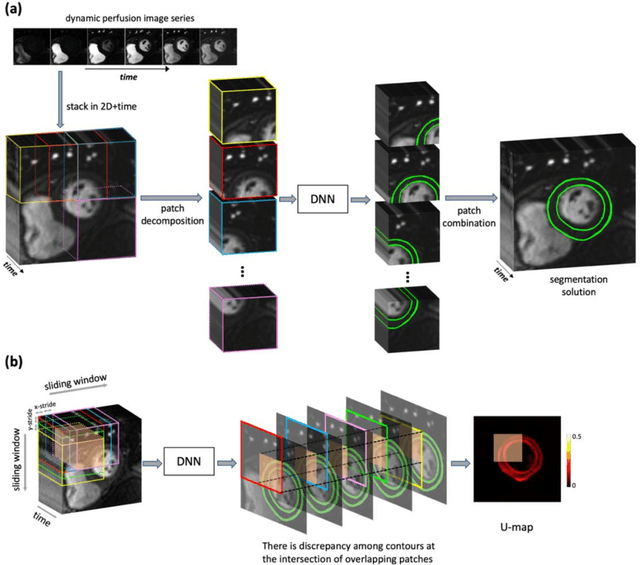
Abstract:Background. Fully automatic analysis of myocardial perfusion MRI datasets enables rapid and objective reporting of stress/rest studies in patients with suspected ischemic heart disease. Developing deep learning techniques that can analyze multi-center datasets despite limited training data and variations in software and hardware is an ongoing challenge. Methods. Datasets from 3 medical centers acquired at 3T (n = 150 subjects) were included: an internal dataset (inD; n = 95) and two external datasets (exDs; n = 55) used for evaluating the robustness of the trained deep neural network (DNN) models against differences in pulse sequence (exD-1) and scanner vendor (exD-2). A subset of inD (n = 85) was used for training/validation of a pool of DNNs for segmentation, all using the same spatiotemporal U-Net architecture and hyperparameters but with different parameter initializations. We employed a space-time sliding-patch analysis approach that automatically yields a pixel-wise "uncertainty map" as a byproduct of the segmentation process. In our approach, a given test case is segmented by all members of the DNN pool and the resulting uncertainty maps are leveraged to automatically select the "best" one among the pool of solutions. Results. The proposed DAUGS analysis approach performed similarly to the established approach on the internal dataset (p = n.s.) whereas it significantly outperformed on the external datasets (p < 0.005 for exD-1 and exD-2). Moreover, the number of image series with "failed" segmentation was significantly lower for the proposed vs. the established approach (4.3% vs. 17.1%, p < 0.0005). Conclusions. The proposed DAUGS analysis approach has the potential to improve the robustness of deep learning methods for segmentation of multi-center stress perfusion datasets with variations in the choice of pulse sequence, site location or scanner vendor.
Data-driven generation of 4D velocity profiles in the aneurysmal ascending aorta
Nov 01, 2022Abstract:Numerical simulations of blood flow are a valuable tool to investigate the pathophysiology of ascending thoracic aortic aneurysms (ATAA). To accurately reproduce hemodynamics, computational fluid dynamics (CFD) models must employ realistic inflow boundary conditions (BCs). However, the limited availability of in vivo velocity measurements still makes researchers resort to idealized BCs. In this study we generated and thoroughly characterized a large dataset of synthetic 4D aortic velocity profiles suitable to be used as BCs for CFD simulations. 4D flow MRI scans of 30 subjects with ATAA were processed to extract cross-sectional planes along the ascending aorta, ensuring spatial alignment among all planes and interpolating all velocity fields to a reference configuration. Velocity profiles of the clinical cohort were extensively characterized by computing flow morphology descriptors of both spatial and temporal features. By exploiting principal component analysis (PCA), a statistical shape model (SSM) of 4D aortic velocity profiles was built and a dataset of 437 synthetic cases with realistic properties was generated. Comparison between clinical and synthetic datasets showed that the synthetic data presented similar characteristics as the clinical population in terms of key morphological parameters. The average velocity profile qualitatively resembled a parabolic-shaped profile, but was quantitatively characterized by more complex flow patterns which an idealized profile would not replicate. Statistically significant correlations were found between PCA principal modes of variation and flow descriptors. We built a data-driven generative model of 4D aortic velocity profiles, suitable to be used in computational studies of blood flow. The proposed software system also allows to map any of the generated velocity profiles to the inlet plane of any virtual subject given its coordinate set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge