Orlando P. Simonetti
Improved Robustness for Deep Learning-based Segmentation of Multi-Center Myocardial Perfusion MRI Datasets Using Data Adaptive Uncertainty-guided Space-time Analysis
Aug 09, 2024


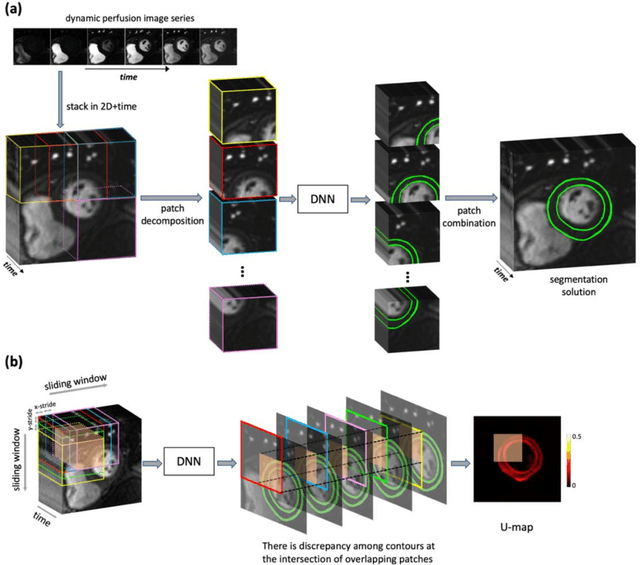
Abstract:Background. Fully automatic analysis of myocardial perfusion MRI datasets enables rapid and objective reporting of stress/rest studies in patients with suspected ischemic heart disease. Developing deep learning techniques that can analyze multi-center datasets despite limited training data and variations in software and hardware is an ongoing challenge. Methods. Datasets from 3 medical centers acquired at 3T (n = 150 subjects) were included: an internal dataset (inD; n = 95) and two external datasets (exDs; n = 55) used for evaluating the robustness of the trained deep neural network (DNN) models against differences in pulse sequence (exD-1) and scanner vendor (exD-2). A subset of inD (n = 85) was used for training/validation of a pool of DNNs for segmentation, all using the same spatiotemporal U-Net architecture and hyperparameters but with different parameter initializations. We employed a space-time sliding-patch analysis approach that automatically yields a pixel-wise "uncertainty map" as a byproduct of the segmentation process. In our approach, a given test case is segmented by all members of the DNN pool and the resulting uncertainty maps are leveraged to automatically select the "best" one among the pool of solutions. Results. The proposed DAUGS analysis approach performed similarly to the established approach on the internal dataset (p = n.s.) whereas it significantly outperformed on the external datasets (p < 0.005 for exD-1 and exD-2). Moreover, the number of image series with "failed" segmentation was significantly lower for the proposed vs. the established approach (4.3% vs. 17.1%, p < 0.0005). Conclusions. The proposed DAUGS analysis approach has the potential to improve the robustness of deep learning methods for segmentation of multi-center stress perfusion datasets with variations in the choice of pulse sequence, site location or scanner vendor.
Cardiac and respiratory motion extraction for MRI using Pilot Tone-a patient study
Jan 31, 2022Abstract:Background:The Pilot Tone (PT) technology allows contactless monitoring of physiological motion during the MRI scan. Several studies have shown that both respiratory and cardiac motion can be extracted from the PT signal successfully. However, most of these studies were performed in healthy volunteers. In this study, we seek to evaluate the accuracy and reliability of the cardiac and respiratory signals extracted from PT in patients clinically referred for cardiovascular MRI (CMR). Methods: Twenty-three patients were included in this study, each scanned under free-breathing conditions using a balanced steady-state free-precession real-time (RT) cine sequence on a 1.5T scanner. The PT signal was generated by a built-in PT transmitter integrated within the body array coil. For comparison, ECG and BioMatrix (BM) respiratory sensor signals were also synchronously recorded. To assess the performances of PT, ECG, and BM, cardiac and respiratory signals extracted from the RT cine images were used as the ground truth. Results: The respiratory motion extracted from PT correlated positively with the image-derived respiratory signal in all cases and showed a stronger correlation (absolute coefficient: 0.95-0.09) than BM (0.72-0.24). For the cardiac signal, the precision of PT-based triggers (standard deviation of PT trigger locations relative to ECG triggers) ranged from 6.6 to 81.2 ms (median 19.5 ms). Overall, the performance of PT-based trigger extraction was comparable to that of ECG. Conclusions: This study demonstrates the potential of PT to monitor both respiratory and cardiac motion in patients clinically referred for CMR.
Identifying Relevant Eigenimages - a Random Matrix Approach
Dec 25, 2008



Abstract:Dimensional reduction of high dimensional data can be achieved by keeping only the relevant eigenmodes after principal component analysis. However, differentiating relevant eigenmodes from the random noise eigenmodes is problematic. A new method based on the random matrix theory and a statistical goodness-of-fit test is proposed in this paper. It is validated by numerical simulations and applied to real-time magnetic resonance cardiac cine images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge