Patryk Rygiel
GReAT: leveraging geometric artery data to improve wall shear stress assessment
Aug 26, 2025Abstract:Leveraging big data for patient care is promising in many medical fields such as cardiovascular health. For example, hemodynamic biomarkers like wall shear stress could be assessed from patient-specific medical images via machine learning algorithms, bypassing the need for time-intensive computational fluid simulation. However, it is extremely challenging to amass large-enough datasets to effectively train such models. We could address this data scarcity by means of self-supervised pre-training and foundations models given large datasets of geometric artery models. In the context of coronary arteries, leveraging learned representations to improve hemodynamic biomarker assessment has not yet been well studied. In this work, we address this gap by investigating whether a large dataset (8449 shapes) consisting of geometric models of 3D blood vessels can benefit wall shear stress assessment in coronary artery models from a small-scale clinical trial (49 patients). We create a self-supervised target for the 3D blood vessels by computing the heat kernel signature, a quantity obtained via Laplacian eigenvectors, which captures the very essence of the shapes. We show how geometric representations learned from this datasets can boost segmentation of coronary arteries into regions of low, mid and high (time-averaged) wall shear stress even when trained on limited data.
Beyond Pixels: Medical Image Quality Assessment with Implicit Neural Representations
Aug 07, 2025Abstract:Artifacts pose a significant challenge in medical imaging, impacting diagnostic accuracy and downstream analysis. While image-based approaches for detecting artifacts can be effective, they often rely on preprocessing methods that can lead to information loss and high-memory-demand medical images, thereby limiting the scalability of classification models. In this work, we propose the use of implicit neural representations (INRs) for image quality assessment. INRs provide a compact and continuous representation of medical images, naturally handling variations in resolution and image size while reducing memory overhead. We develop deep weight space networks, graph neural networks, and relational attention transformers that operate on INRs to achieve image quality assessment. Our method is evaluated on the ACDC dataset with synthetically generated artifact patterns, demonstrating its effectiveness in assessing image quality while achieving similar performance with fewer parameters.
Wall Shear Stress Estimation in Abdominal Aortic Aneurysms: Towards Generalisable Neural Surrogate Models
Jul 30, 2025Abstract:Abdominal aortic aneurysms (AAAs) are pathologic dilatations of the abdominal aorta posing a high fatality risk upon rupture. Studying AAA progression and rupture risk often involves in-silico blood flow modelling with computational fluid dynamics (CFD) and extraction of hemodynamic factors like time-averaged wall shear stress (TAWSS) or oscillatory shear index (OSI). However, CFD simulations are known to be computationally demanding. Hence, in recent years, geometric deep learning methods, operating directly on 3D shapes, have been proposed as compelling surrogates, estimating hemodynamic parameters in just a few seconds. In this work, we propose a geometric deep learning approach to estimating hemodynamics in AAA patients, and study its generalisability to common factors of real-world variation. We propose an E(3)-equivariant deep learning model utilising novel robust geometrical descriptors and projective geometric algebra. Our model is trained to estimate transient WSS using a dataset of CT scans of 100 AAA patients, from which lumen geometries are extracted and reference CFD simulations with varying boundary conditions are obtained. Results show that the model generalizes well within the distribution, as well as to the external test set. Moreover, the model can accurately estimate hemodynamics across geometry remodelling and changes in boundary conditions. Furthermore, we find that a trained model can be applied to different artery tree topologies, where new and unseen branches are added during inference. Finally, we find that the model is to a large extent agnostic to mesh resolution. These results show the accuracy and generalisation of the proposed model, and highlight its potential to contribute to hemodynamic parameter estimation in clinical practice.
Geometric deep learning for local growth prediction on abdominal aortic aneurysm surfaces
Jun 11, 2025Abstract:Abdominal aortic aneurysms (AAAs) are progressive focal dilatations of the abdominal aorta. AAAs may rupture, with a survival rate of only 20\%. Current clinical guidelines recommend elective surgical repair when the maximum AAA diameter exceeds 55 mm in men or 50 mm in women. Patients that do not meet these criteria are periodically monitored, with surveillance intervals based on the maximum AAA diameter. However, this diameter does not take into account the complex relation between the 3D AAA shape and its growth, making standardized intervals potentially unfit. Personalized AAA growth predictions could improve monitoring strategies. We propose to use an SE(3)-symmetric transformer model to predict AAA growth directly on the vascular model surface enriched with local, multi-physical features. In contrast to other works which have parameterized the AAA shape, this representation preserves the vascular surface's anatomical structure and geometric fidelity. We train our model using a longitudinal dataset of 113 computed tomography angiography (CTA) scans of 24 AAA patients at irregularly sampled intervals. After training, our model predicts AAA growth to the next scan moment with a median diameter error of 1.18 mm. We further demonstrate our model's utility to identify whether a patient will become eligible for elective repair within two years (acc = 0.93). Finally, we evaluate our model's generalization on an external validation set consisting of 25 CTAs from 7 AAA patients from a different hospital. Our results show that local directional AAA growth prediction from the vascular surface is feasible and may contribute to personalized surveillance strategies.
Active Learning for Deep Learning-Based Hemodynamic Parameter Estimation
Mar 05, 2025Abstract:Hemodynamic parameters such as pressure and wall shear stress play an important role in diagnosis, prognosis, and treatment planning in cardiovascular diseases. These parameters can be accurately computed using computational fluid dynamics (CFD), but CFD is computationally intensive. Hence, deep learning methods have been adopted as a surrogate to rapidly estimate CFD outcomes. A drawback of such data-driven models is the need for time-consuming reference CFD simulations for training. In this work, we introduce an active learning framework to reduce the number of CFD simulations required for the training of surrogate models, lowering the barriers to their deployment in new applications. We propose three distinct querying strategies to determine for which unlabeled samples CFD simulations should be obtained. These querying strategies are based on geometrical variance, ensemble uncertainty, and adherence to the physics governing fluid dynamics. We benchmark these methods on velocity field estimation in synthetic coronary artery bifurcations and find that they allow for substantial reductions in annotation cost. Notably, we find that our strategies reduce the number of samples required by up to 50% and make the trained models more robust to difficult cases. Our results show that active learning is a feasible strategy to increase the potential of deep learning-based CFD surrogates.
Learning Hemodynamic Scalar Fields on Coronary Artery Meshes: A Benchmark of Geometric Deep Learning Models
Jan 15, 2025



Abstract:Coronary artery disease, caused by the narrowing of coronary vessels due to atherosclerosis, is the leading cause of death worldwide. The diagnostic gold standard, fractional flow reserve (FFR), measures the trans-stenotic pressure ratio during maximal vasodilation but is invasive and costly. This has driven the development of virtual FFR (vFFR) using computational fluid dynamics (CFD) to simulate coronary flow. Geometric deep learning algorithms have shown promise for learning features on meshes, including cardiovascular research applications. This study empirically analyzes various backends for predicting vFFR fields in coronary arteries as CFD surrogates, comparing six backends for learning hemodynamics on meshes using CFD solutions as ground truth. The study has two parts: i) Using 1,500 synthetic left coronary artery bifurcations, models were trained to predict pressure-related fields for vFFR reconstruction, comparing different learning variables. ii) Using 427 patient-specific CFD simulations, experiments were repeated focusing on the best-performing learning variable from the synthetic dataset. Most backends performed well on the synthetic dataset, especially when predicting pressure drop over the manifold. Transformer-based backends outperformed others when predicting pressure and vFFR fields and were the only models achieving strong performance on patient-specific data, excelling in both average per-point error and vFFR accuracy in stenotic lesions. These results suggest geometric deep learning backends can effectively replace CFD for simple geometries, while transformer-based networks are superior for complex, heterogeneous datasets. Pressure drop was identified as the optimal network output for learning pressure-related fields.
Deep vectorised operators for pulsatile hemodynamics estimation in coronary arteries from a steady-state prior
Oct 15, 2024



Abstract:Cardiovascular hemodynamic fields provide valuable medical decision markers for coronary artery disease. Computational fluid dynamics (CFD) is the gold standard for accurate, non-invasive evaluation of these quantities in vivo. In this work, we propose a time-efficient surrogate model, powered by machine learning, for the estimation of pulsatile hemodynamics based on steady-state priors. We introduce deep vectorised operators, a modelling framework for discretisation independent learning on infinite-dimensional function spaces. The underlying neural architecture is a neural field conditioned on hemodynamic boundary conditions. Importantly, we show how relaxing the requirement of point-wise action to permutation-equivariance leads to a family of models that can be parametrised by message passing and self-attention layers. We evaluate our approach on a dataset of 74 stenotic coronary arteries extracted from coronary computed tomography angiography (CCTA) with patient-specific pulsatile CFD simulations as ground truth. We show that our model produces accurate estimates of the pulsatile velocity and pressure while being agnostic to re-sampling of the source domain (discretisation independence). This shows that deep vectorised operators are a powerful modelling tool for cardiovascular hemodynamics estimation in coronary arteries and beyond.
ICML Topological Deep Learning Challenge 2024: Beyond the Graph Domain
Sep 08, 2024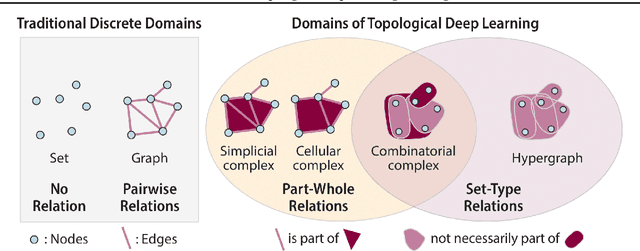
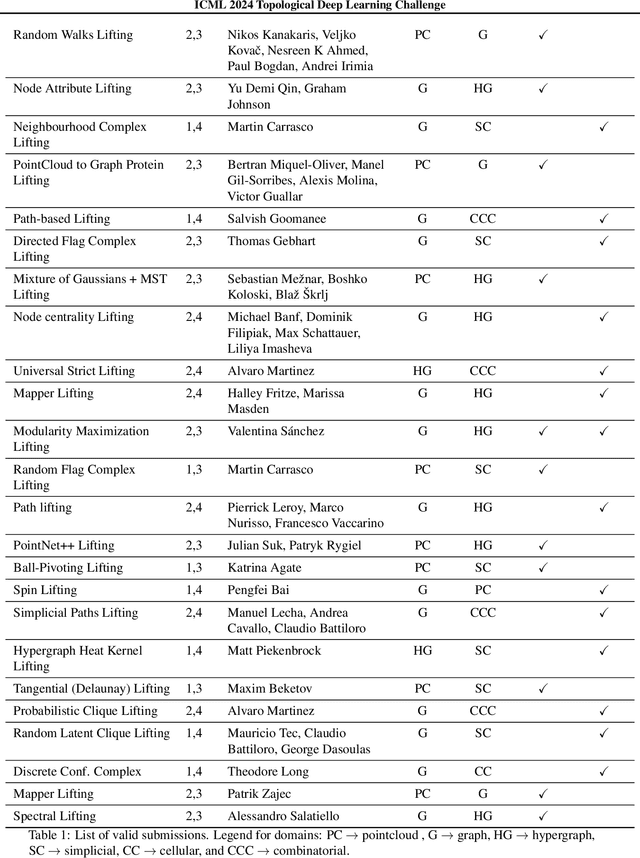
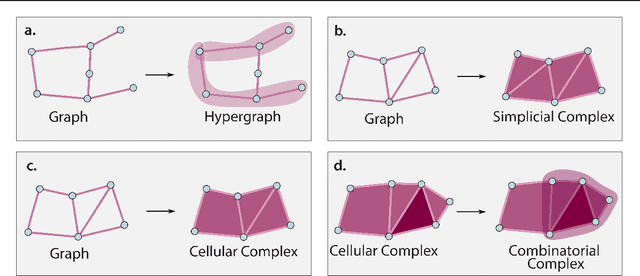
Abstract:This paper describes the 2nd edition of the ICML Topological Deep Learning Challenge that was hosted within the ICML 2024 ELLIS Workshop on Geometry-grounded Representation Learning and Generative Modeling (GRaM). The challenge focused on the problem of representing data in different discrete topological domains in order to bridge the gap between Topological Deep Learning (TDL) and other types of structured datasets (e.g. point clouds, graphs). Specifically, participants were asked to design and implement topological liftings, i.e. mappings between different data structures and topological domains --like hypergraphs, or simplicial/cell/combinatorial complexes. The challenge received 52 submissions satisfying all the requirements. This paper introduces the main scope of the challenge, and summarizes the main results and findings.
Neural Fields for Continuous Periodic Motion Estimation in 4D Cardiovascular Imaging
Jul 30, 2024



Abstract:Time-resolved three-dimensional flow MRI (4D flow MRI) provides a unique non-invasive solution to visualize and quantify hemodynamics in blood vessels such as the aortic arch. However, most current analysis methods for arterial 4D flow MRI use static artery walls because of the difficulty in obtaining a full cycle segmentation. To overcome this limitation, we propose a neural fields-based method that directly estimates continuous periodic wall deformations throughout the cardiac cycle. For a 3D + time imaging dataset, we optimize an implicit neural representation (INR) that represents a time-dependent velocity vector field (VVF). An ODE solver is used to integrate the VVF into a deformation vector field (DVF), that can deform images, segmentation masks, or meshes over time, thereby visualizing and quantifying local wall motion patterns. To properly reflect the periodic nature of 3D + time cardiovascular data, we impose periodicity in two ways. First, by periodically encoding the time input to the INR, and hence VVF. Second, by regularizing the DVF. We demonstrate the effectiveness of this approach on synthetic data with different periodic patterns, ECG-gated CT, and 4D flow MRI data. The obtained method could be used to improve 4D flow MRI analysis.
Global Control for Local SO-Equivariant Scale-Invariant Vessel Segmentation
Mar 22, 2024



Abstract:Personalized 3D vascular models can aid in a range of diagnostic, prognostic, and treatment-planning tasks relevant to cardiovascular disease management. Deep learning provides a means to automatically obtain such models. Ideally, a user should have control over the exact region of interest (ROI) to be included in a vascular model, and the model should be watertight and highly accurate. To this end, we propose a combination of a global controller leveraging voxel mask segmentations to provide boundary conditions for vessels of interest to a local, iterative vessel segmentation model. We introduce the preservation of scale- and rotational symmetries in the local segmentation model, leading to generalisation to vessels of unseen sizes and orientations. Combined with the global controller, this enables flexible 3D vascular model building, without additional retraining. We demonstrate the potential of our method on a dataset containing abdominal aortic aneurysms (AAAs). Our method performs on par with a state-of-the-art segmentation model in the segmentation of AAAs, iliac arteries and renal arteries, while providing a watertight, smooth surface segmentation. Moreover, we demonstrate that by adapting the global controller, we can easily extend vessel sections in the 3D model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge