Riccardo Bellazzi
Exploring the Impact of Environmental Pollutants on Multiple Sclerosis Progression
Aug 30, 2024


Abstract:Multiple Sclerosis (MS) is a chronic autoimmune and inflammatory neurological disorder characterised by episodes of symptom exacerbation, known as relapses. In this study, we investigate the role of environmental factors in relapse occurrence among MS patients, using data from the H2020 BRAINTEASER project. We employed predictive models, including Random Forest (RF) and Logistic Regression (LR), with varying sets of input features to predict the occurrence of relapses based on clinical and pollutant data collected over a week. The RF yielded the best result, with an AUC-ROC score of 0.713. Environmental variables, such as precipitation, NO2, PM2.5, humidity, and temperature, were found to be relevant to the prediction.
Reshaping Free-Text Radiology Notes Into Structured Reports With Generative Transformers
Mar 27, 2024Abstract:BACKGROUND: Radiology reports are typically written in a free-text format, making clinical information difficult to extract and use. Recently the adoption of structured reporting (SR) has been recommended by various medical societies thanks to the advantages it offers, e.g. standardization, completeness and information retrieval. We propose a pipeline to extract information from free-text radiology reports, that fits with the items of the reference SR registry proposed by a national society of interventional and medical radiology, focusing on CT staging of patients with lymphoma. METHODS: Our work aims to leverage the potential of Natural Language Processing (NLP) and Transformer-based models to deal with automatic SR registry filling. With the availability of 174 radiology reports, we investigate a rule-free generative Question Answering approach based on a domain-specific version of T5 (IT5). Two strategies (batch-truncation and ex-post combination) are implemented to comply with the model's context length limitations. Performance is evaluated in terms of strict accuracy, F1, and format accuracy, and compared with the widely used GPT-3.5 Large Language Model. A 5-point Likert scale questionnaire is used to collect human-expert feedback on the similarity between medical annotations and generated answers. RESULTS: The combination of fine-tuning and batch splitting allows IT5 to achieve notable results; it performs on par with GPT-3.5 albeit its size being a thousand times smaller in terms of parameters. Human-based assessment scores show a high correlation (Spearman's correlation coefficients>0.88, p-values<0.001) with AI performance metrics (F1) and confirm the superior ability of LLMs (i.e., GPT-3.5, 175B of parameters) in generating plausible human-like statements.
Evaluation of Predictive Reliability to Foster Trust in Artificial Intelligence. A case study in Multiple Sclerosis
Feb 27, 2024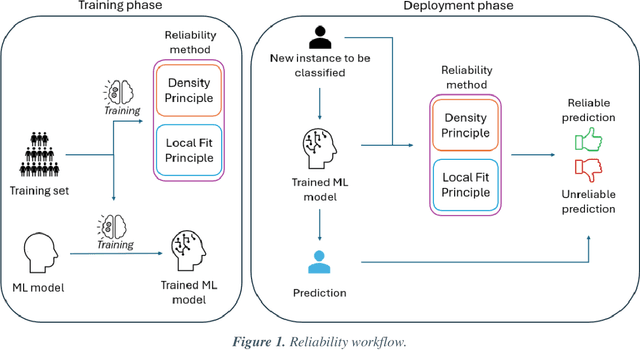
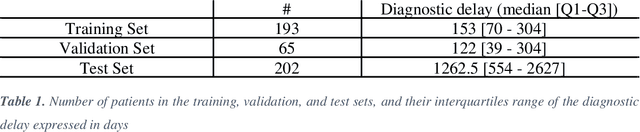
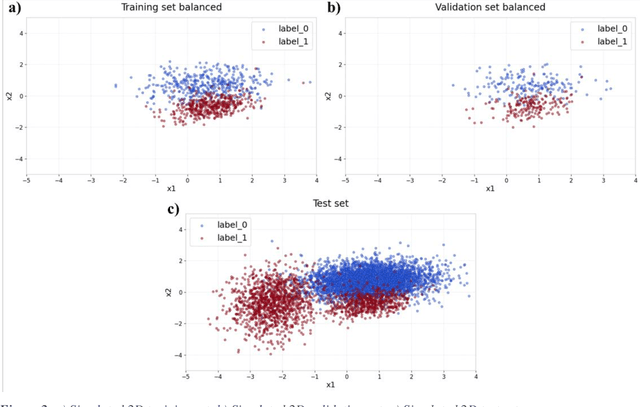
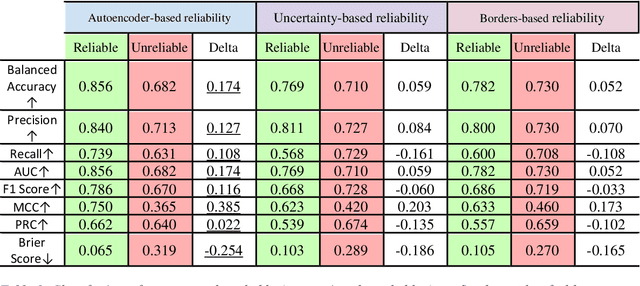
Abstract:Applying Artificial Intelligence (AI) and Machine Learning (ML) in critical contexts, such as medicine, requires the implementation of safety measures to reduce risks of harm in case of prediction errors. Spotting ML failures is of paramount importance when ML predictions are used to drive clinical decisions. ML predictive reliability measures the degree of trust of a ML prediction on a new instance, thus allowing decision-makers to accept or reject it based on its reliability. To assess reliability, we propose a method that implements two principles. First, our approach evaluates whether an instance to be classified is coming from the same distribution of the training set. To do this, we leverage Autoencoders (AEs) ability to reconstruct the training set with low error. An instance is considered Out-of-Distribution (OOD) if the AE reconstructs it with a high error. Second, it is evaluated whether the ML classifier has good performances on samples similar to the newly classified instance by using a proxy model. We show that this approach is able to assess reliability both in a simulated scenario and on a model trained to predict disease progression of Multiple Sclerosis patients. We also developed a Python package, named relAI, to embed reliability measures into ML pipelines. We propose a simple approach that can be used in the deployment phase of any ML model to suggest whether to trust predictions or not. Our method holds the promise to provide effective support to clinicians by spotting potential ML failures during deployment.
Advancing Italian Biomedical Information Extraction with Large Language Models: Methodological Insights and Multicenter Practical Application
Jun 08, 2023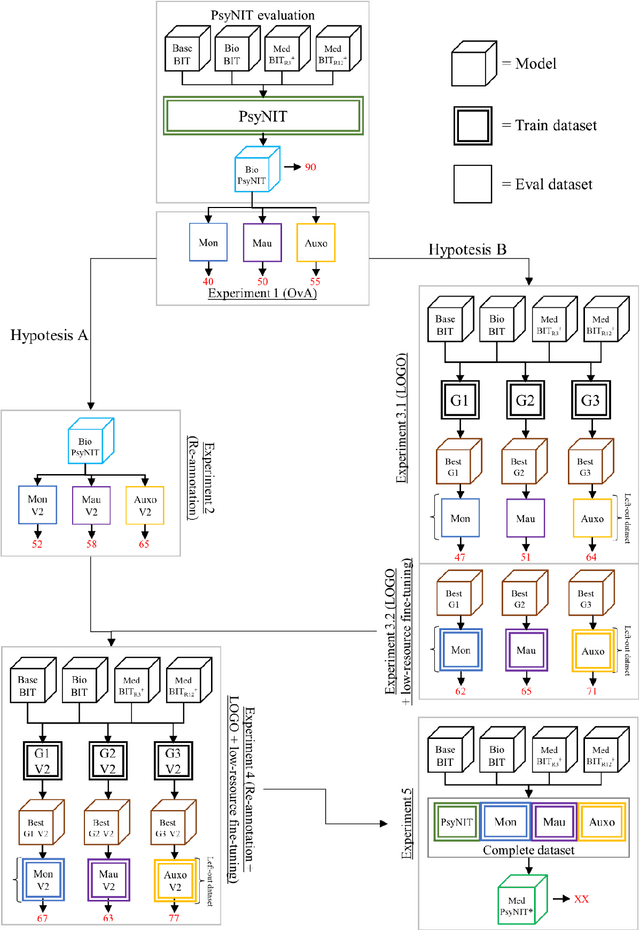


Abstract:The introduction of computerized medical records in hospitals has reduced burdensome operations like manual writing and information fetching. However, the data contained in medical records are still far underutilized, primarily because extracting them from unstructured textual medical records takes time and effort. Information Extraction, a subfield of Natural Language Processing, can help clinical practitioners overcome this limitation, using automated text-mining pipelines. In this work, we created the first Italian neuropsychiatric Named Entity Recognition dataset, PsyNIT, and used it to develop a Large Language Model for this task. Moreover, we conducted several experiments with three external independent datasets to implement an effective multicenter model, with overall F1-score 84.77%, Precision 83.16%, Recall 86.44%. The lessons learned are: (i) the crucial role of a consistent annotation process and (ii) a fine-tuning strategy that combines classical methods with a "few-shot" approach. This allowed us to establish methodological guidelines that pave the way for future implementations in this field and allow Italian hospitals to tap into important research opportunities.
CAD-RADS scoring of coronary CT angiography with Multi-Axis Vision Transformer: a clinically-inspired deep learning pipeline
Apr 14, 2023Abstract:The standard non-invasive imaging technique used to assess the severity and extent of Coronary Artery Disease (CAD) is Coronary Computed Tomography Angiography (CCTA). However, manual grading of each patient's CCTA according to the CAD-Reporting and Data System (CAD-RADS) scoring is time-consuming and operator-dependent, especially in borderline cases. This work proposes a fully automated, and visually explainable, deep learning pipeline to be used as a decision support system for the CAD screening procedure. The pipeline performs two classification tasks: firstly, identifying patients who require further clinical investigations and secondly, classifying patients into subgroups based on the degree of stenosis, according to commonly used CAD-RADS thresholds. The pipeline pre-processes multiplanar projections of the coronary arteries, extracted from the original CCTAs, and classifies them using a fine-tuned Multi-Axis Vision Transformer architecture. With the aim of emulating the current clinical practice, the model is trained to assign a per-patient score by stacking the bi-dimensional longitudinal cross-sections of the three main coronary arteries along channel dimension. Furthermore, it generates visually interpretable maps to assess the reliability of the predictions. When run on a database of 1873 three-channel images of 253 patients collected at the Monzino Cardiology Center in Milan, the pipeline obtained an AUC of 0.87 and 0.93 for the two classification tasks, respectively. According to our knowledge, this is the first model trained to assign CAD-RADS scores learning solely from patient scores and not requiring finer imaging annotation steps that are not part of the clinical routine.
Localising In-Domain Adaptation of Transformer-Based Biomedical Language Models
Dec 22, 2022Abstract:In the era of digital healthcare, the huge volumes of textual information generated every day in hospitals constitute an essential but underused asset that could be exploited with task-specific, fine-tuned biomedical language representation models, improving patient care and management. For such specialized domains, previous research has shown that fine-tuning models stemming from broad-coverage checkpoints can largely benefit additional training rounds over large-scale in-domain resources. However, these resources are often unreachable for less-resourced languages like Italian, preventing local medical institutions to employ in-domain adaptation. In order to reduce this gap, our work investigates two accessible approaches to derive biomedical language models in languages other than English, taking Italian as a concrete use-case: one based on neural machine translation of English resources, favoring quantity over quality; the other based on a high-grade, narrow-scoped corpus natively written in Italian, thus preferring quality over quantity. Our study shows that data quantity is a harder constraint than data quality for biomedical adaptation, but the concatenation of high-quality data can improve model performance even when dealing with relatively size-limited corpora. The models published from our investigations have the potential to unlock important research opportunities for Italian hospitals and academia. Finally, the set of lessons learned from the study constitutes valuable insights towards a solution to build biomedical language models that are generalizable to other less-resourced languages and different domain settings.
Tree-based local explanations of machine learning model predictions, AraucanaXAI
Oct 15, 2021Abstract:Increasingly complex learning methods such as boosting, bagging and deep learning have made ML models more accurate, but harder to understand and interpret. A tradeoff between performance and intelligibility is often to be faced, especially in high-stakes applications like medicine. In the present article we propose a novel methodological approach for generating explanations of the predictions of a generic ML model, given a specific instance for which the prediction has been made, that can tackle both classification and regression tasks. Advantages of the proposed XAI approach include improved fidelity to the original model, the ability to deal with non-linear decision boundaries, and native support to both classification and regression problems
The Gene Mover's Distance: Single-cell similarity via Optimal Transport
Feb 01, 2021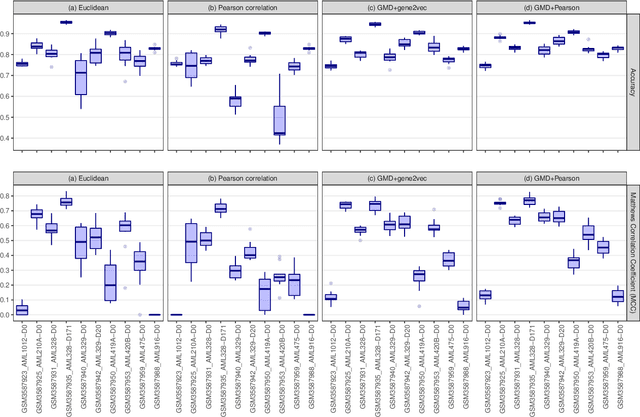
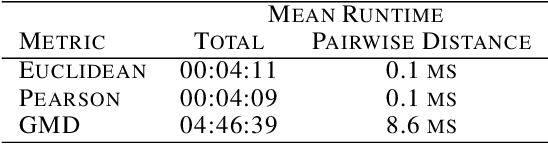
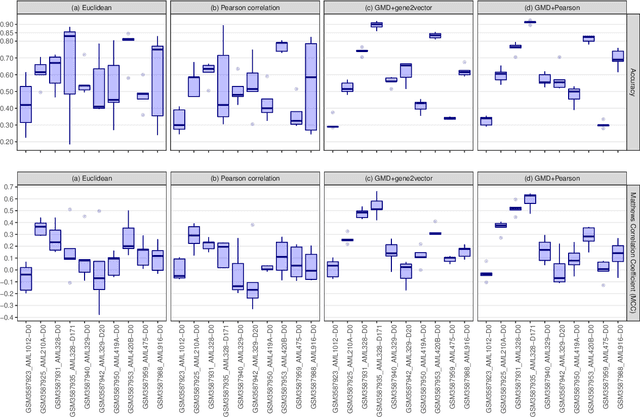
Abstract:This paper introduces the Gene Mover's Distance, a measure of similarity between a pair of cells based on their gene expression profiles obtained via single-cell RNA sequencing. The underlying idea of the proposed distance is to interpret the gene expression array of a single cell as a discrete probability measure. The distance between two cells is hence computed by solving an Optimal Transport problem between the two corresponding discrete measures. In the Optimal Transport model, we use two types of cost function for measuring the distance between a pair of genes. The first cost function exploits a gene embedding, called gene2vec, which is used to map each gene to a high dimensional vector: the cost of moving a unit of mass of gene expression from a gene to another is set to the Euclidean distance between the corresponding embedded vectors. The second cost function is based on a Pearson distance among pairs of genes. In both cost functions, the more two genes are correlated, the lower is their distance. We exploit the Gene Mover's Distance to solve two classification problems: the classification of cells according to their condition and according to their type. To assess the impact of our new metric, we compare the performances of a $k$-Nearest Neighbor classifier using different distances. The computational results show that the Gene Mover's Distance is competitive with the state-of-the-art distances used in the literature.
Temporal Reasoning with Probabilities
Mar 27, 2013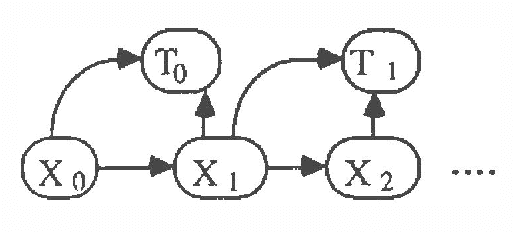
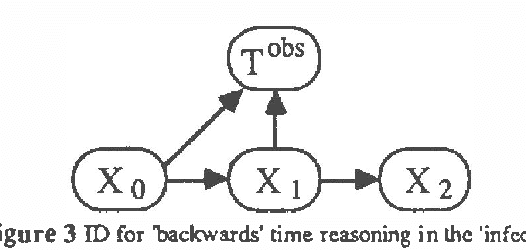

Abstract:In this paper we explore representations of temporal knowledge based upon the formalism of Causal Probabilistic Networks (CPNs). Two different ?continuous-time? representations are proposed. In the first, the CPN includes variables representing ?event-occurrence times?, possibly on different time scales, and variables representing the ?state? of the system at these times. In the second, the CPN describes the influences between random variables with values in () representing dates, i.e. time-points associated with the occurrence of relevant events. However, structuring a system of inter-related dates as a network where all links commit to a single specific notion of cause and effect is in general far from trivial and leads to severe difficulties. We claim that we should recognize explicitly different kinds of relation between dates, such as ?cause?, ?inhibition?, ?competition?, etc., and propose a method whereby these relations are coherently embedded in a CPN using additional auxiliary nodes corresponding to "instrumental" variables. Also discussed, though not covered in detail, is the topic concerning how the quantitative specifications to be inserted in a temporal CPN can be learned from specific data.
Bayesian Networks Aplied to Therapy Monitoring
Mar 20, 2013



Abstract:We propose a general Bayesian network model for application in a wide class of problems of therapy monitoring. We discuss the use of stochastic simulation as a computational approach to inference on the proposed class of models. As an illustration we present an application to the monitoring of cytotoxic chemotherapy in breast cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge