Andrea Codegoni
Open-CD: A Comprehensive Toolbox for Change Detection
Jul 22, 2024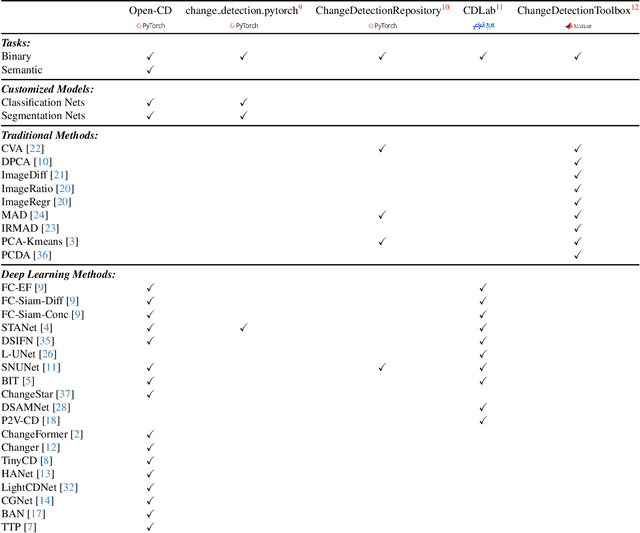
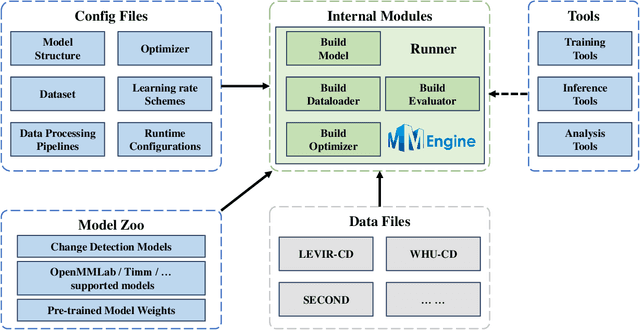
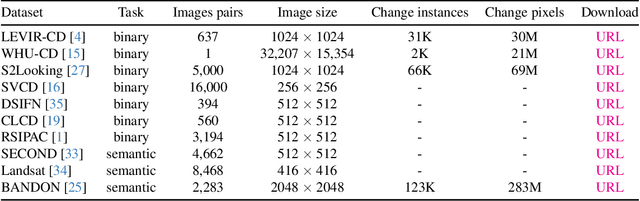
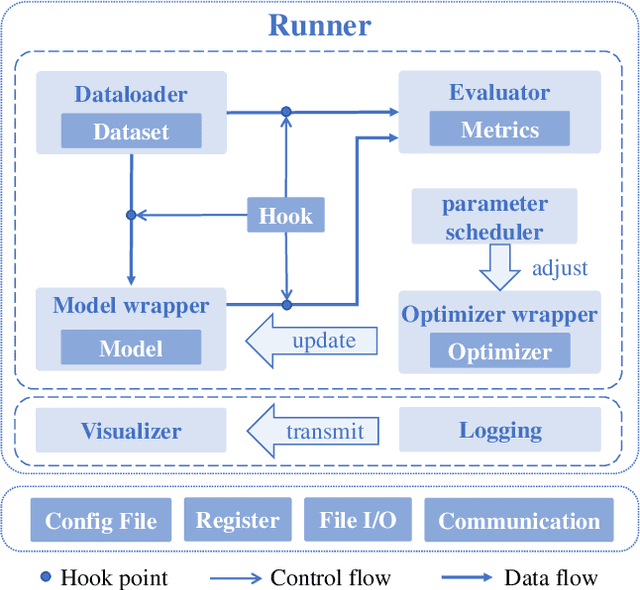
Abstract:We present Open-CD, a change detection toolbox that contains a rich set of change detection methods as well as related components and modules. The toolbox started from a series of open source general vision task tools, including OpenMMLab Toolkits, PyTorch Image Models, etc. It gradually evolves into a unified platform that covers many popular change detection methods and contemporary modules. It not only includes training and inference codes, but also provides some useful scripts for data analysis. We believe this toolbox is by far the most complete change detection toolbox. In this report, we introduce the various features, supported methods and applications of Open-CD. In addition, we also conduct a benchmarking study on different methods and components. We wish that the toolbox and benchmark could serve the growing research community by providing a flexible toolkit to reimplement existing methods and develop their own new change detectors. Code and models are available at \url{https://github.com/likyoo/open-cd}. Pioneeringly, this report also includes brief descriptions of the algorithms supported in Open-CD, mainly contributed by their authors. We sincerely encourage researchers in this field to participate in this project and work together to create a more open community. This toolkit and report will be kept updated.
TINYCD: A Deep Learning Model For Change Detection
Jul 26, 2022



Abstract:The aim of change detection (CD) is to detect changes occurred in the same area by comparing two images of that place taken at different times. The challenging part of the CD is to keep track of the changes the user wants to highlight, such as new buildings, and to ignore changes due to external factors such as environmental, lighting condition, fog or seasonal changes. Recent developments in the field of deep learning enabled researchers to achieve outstanding performance in this area. In particular, different mechanisms of space-time attention allowed to exploit the spatial features that are extracted from the models and to correlate them also in a temporal way by exploiting both the available images. The downside is that the models have become increasingly complex and large, often unfeasible for edge applications. These are limitations when the models must be applied to the industrial field or in applications requiring real-time performances. In this work we propose a novel model, called TinyCD, demonstrating to be both lightweight and effective, able to achieve performances comparable or even superior to the current state of the art with 13-150X fewer parameters. In our approach we have exploited the importance of low-level features to compare images. To do this, we use only few backbone blocks. This strategy allow us to keep the number of network parameters low. To compose the features extracted from the two images, we introduce a novel, economical in terms of parameters, mixing block capable of cross correlating features in both space and time domains. Finally, to fully exploit the information contained in the computed features, we define the PW-MLP block able to perform a pixel wise classification. Source code, models and results are available here: https://github.com/AndreaCodegoni/Tiny_model_4_CD
The Gene Mover's Distance: Single-cell similarity via Optimal Transport
Feb 01, 2021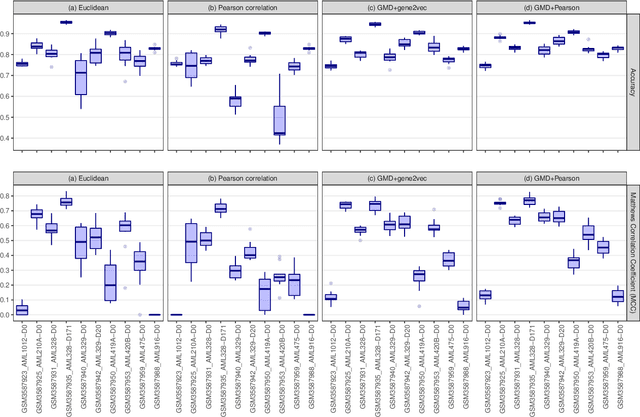
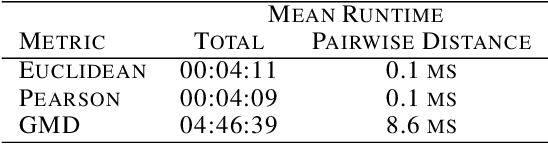
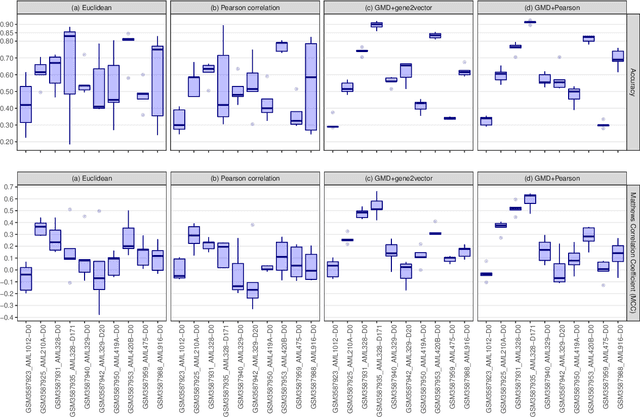
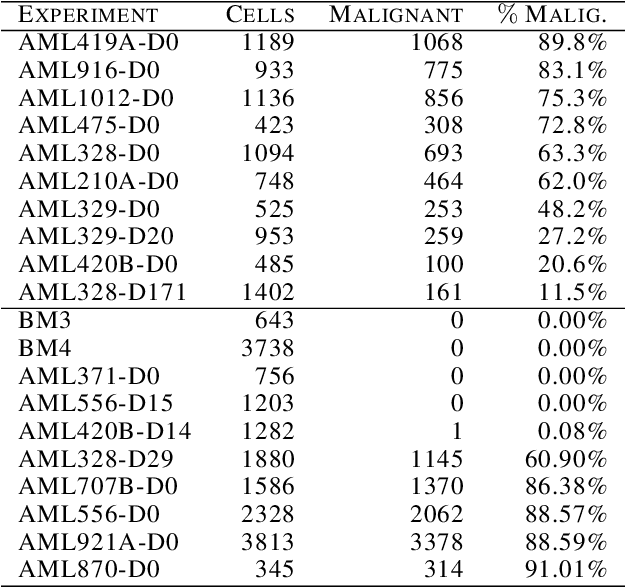
Abstract:This paper introduces the Gene Mover's Distance, a measure of similarity between a pair of cells based on their gene expression profiles obtained via single-cell RNA sequencing. The underlying idea of the proposed distance is to interpret the gene expression array of a single cell as a discrete probability measure. The distance between two cells is hence computed by solving an Optimal Transport problem between the two corresponding discrete measures. In the Optimal Transport model, we use two types of cost function for measuring the distance between a pair of genes. The first cost function exploits a gene embedding, called gene2vec, which is used to map each gene to a high dimensional vector: the cost of moving a unit of mass of gene expression from a gene to another is set to the Euclidean distance between the corresponding embedded vectors. The second cost function is based on a Pearson distance among pairs of genes. In both cost functions, the more two genes are correlated, the lower is their distance. We exploit the Gene Mover's Distance to solve two classification problems: the classification of cells according to their condition and according to their type. To assess the impact of our new metric, we compare the performances of a $k$-Nearest Neighbor classifier using different distances. The computational results show that the Gene Mover's Distance is competitive with the state-of-the-art distances used in the literature.
The Equivalence of Fourier-based and Wasserstein Metrics on Imaging Problems
May 13, 2020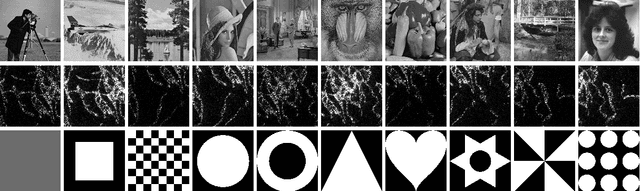
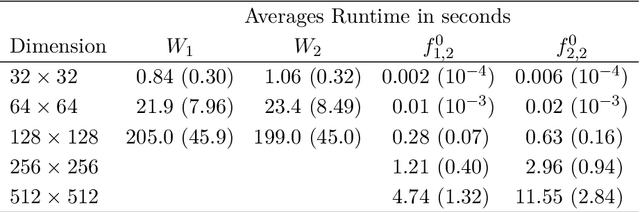
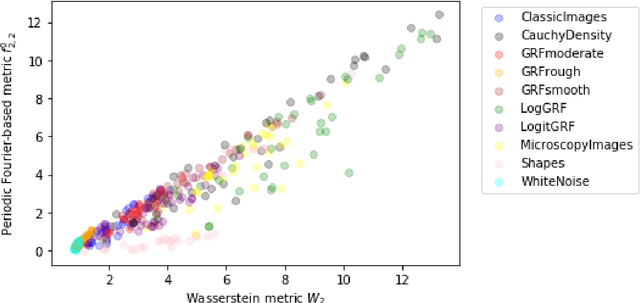
Abstract:We investigate properties of some extensions of a class of Fourier-based probability metrics, originally introduced to study convergence to equilibrium for the solution to the spatially homogeneous Boltzmann equation. At difference with the original one, the new Fourier-based metrics are well-defined also for probability distributions with different centers of mass, and for discrete probability measures supported over a regular grid. Among other properties, it is shown that, in the discrete setting, these new Fourier-based metrics are equivalent either to the Euclidean-Wasserstein distance $W_2$, or to the Kantorovich-Wasserstein distance $W_1$, with explicit constants of equivalence. Numerical results then show that in benchmark problems of image processing, Fourier metrics provide a better runtime with respect to Wasserstein ones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge