Enea Parimbelli
RAR: Setting Knowledge Tripwires for Retrieval Augmented Rejection
May 19, 2025Abstract:Content moderation for large language models (LLMs) remains a significant challenge, requiring flexible and adaptable solutions that can quickly respond to emerging threats. This paper introduces Retrieval Augmented Rejection (RAR), a novel approach that leverages a retrieval-augmented generation (RAG) architecture to dynamically reject unsafe user queries without model retraining. By strategically inserting and marking malicious documents into the vector database, the system can identify and reject harmful requests when these documents are retrieved. Our preliminary results show that RAR achieves comparable performance to embedded moderation in LLMs like Claude 3.5 Sonnet, while offering superior flexibility and real-time customization capabilities, a fundamental feature to timely address critical vulnerabilities. This approach introduces no architectural changes to existing RAG systems, requiring only the addition of specially crafted documents and a simple rejection mechanism based on retrieval results.
Introducing δ-XAI: a novel sensitivity-based method for local AI explanations
Jul 29, 2024
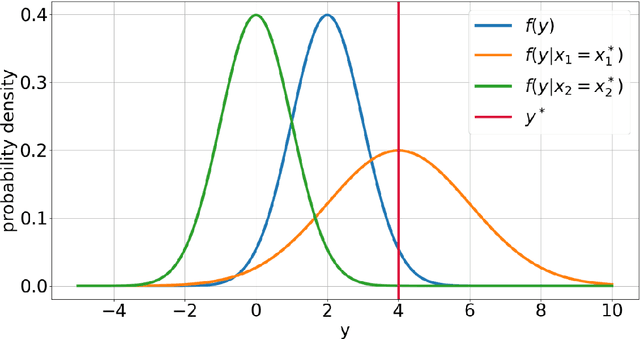

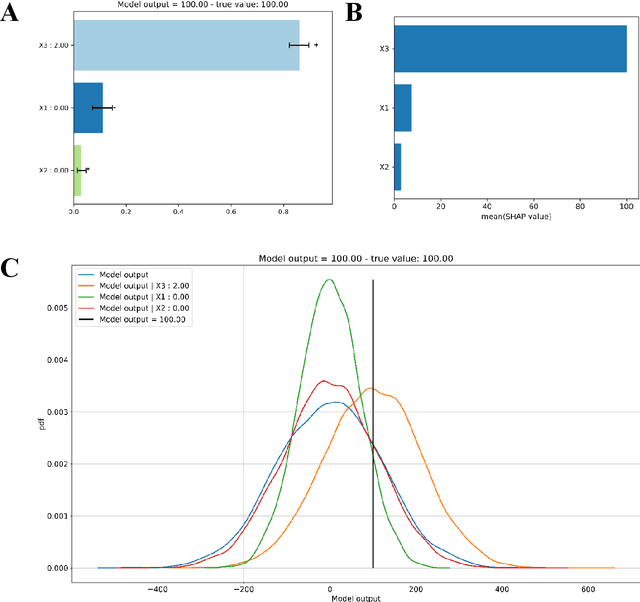
Abstract:Explainable Artificial Intelligence (XAI) is central to the debate on integrating Artificial Intelligence (AI) and Machine Learning (ML) algorithms into clinical practice. High-performing AI/ML models, such as ensemble learners and deep neural networks, often lack interpretability, hampering clinicians' trust in their predictions. To address this, XAI techniques are being developed to describe AI/ML predictions in human-understandable terms. One promising direction is the adaptation of sensitivity analysis (SA) and global sensitivity analysis (GSA), which inherently rank model inputs by their impact on predictions. Here, we introduce a novel delta-XAI method that provides local explanations of ML model predictions by extending the delta index, a GSA metric. The delta-XAI index assesses the impact of each feature's value on the predicted output for individual instances in both regression and classification problems. We formalize the delta-XAI index and provide code for its implementation. The delta-XAI method was evaluated on simulated scenarios using linear regression models, with Shapley values serving as a benchmark. Results showed that the delta-XAI index is generally consistent with Shapley values, with notable discrepancies in models with highly impactful or extreme feature values. The delta-XAI index demonstrated higher sensitivity in detecting dominant features and handling extreme feature values. Qualitatively, the delta-XAI provides intuitive explanations by leveraging probability density functions, making feature rankings clearer and more explainable for practitioners. Overall, the delta-XAI method appears promising for robustly obtaining local explanations of ML model predictions. Further investigations in real-world clinical settings will be conducted to evaluate its impact on AI-assisted clinical workflows.
Igea: a Decoder-Only Language Model for Biomedical Text Generation in Italian
Jul 08, 2024



Abstract:The development of domain-specific language models has significantly advanced natural language processing applications in various specialized fields, particularly in biomedicine. However, the focus has largely been on English-language models, leaving a gap for less-resourced languages such as Italian. This paper introduces Igea, the first decoder-only language model designed explicitly for biomedical text generation in Italian. Built on the Minerva model and continually pretrained on a diverse corpus of Italian medical texts, Igea is available in three model sizes: 350 million, 1 billion, and 3 billion parameters. The models aim to balance computational efficiency and performance, addressing the challenges of managing the peculiarities of medical terminology in Italian. We evaluate Igea using a mix of in-domain biomedical corpora and general-purpose benchmarks, highlighting its efficacy and retention of general knowledge even after the domain-specific training. This paper discusses the model's development and evaluation, providing a foundation for future advancements in Italian biomedical NLP.
Reshaping Free-Text Radiology Notes Into Structured Reports With Generative Transformers
Mar 27, 2024Abstract:BACKGROUND: Radiology reports are typically written in a free-text format, making clinical information difficult to extract and use. Recently the adoption of structured reporting (SR) has been recommended by various medical societies thanks to the advantages it offers, e.g. standardization, completeness and information retrieval. We propose a pipeline to extract information from free-text radiology reports, that fits with the items of the reference SR registry proposed by a national society of interventional and medical radiology, focusing on CT staging of patients with lymphoma. METHODS: Our work aims to leverage the potential of Natural Language Processing (NLP) and Transformer-based models to deal with automatic SR registry filling. With the availability of 174 radiology reports, we investigate a rule-free generative Question Answering approach based on a domain-specific version of T5 (IT5). Two strategies (batch-truncation and ex-post combination) are implemented to comply with the model's context length limitations. Performance is evaluated in terms of strict accuracy, F1, and format accuracy, and compared with the widely used GPT-3.5 Large Language Model. A 5-point Likert scale questionnaire is used to collect human-expert feedback on the similarity between medical annotations and generated answers. RESULTS: The combination of fine-tuning and batch splitting allows IT5 to achieve notable results; it performs on par with GPT-3.5 albeit its size being a thousand times smaller in terms of parameters. Human-based assessment scores show a high correlation (Spearman's correlation coefficients>0.88, p-values<0.001) with AI performance metrics (F1) and confirm the superior ability of LLMs (i.e., GPT-3.5, 175B of parameters) in generating plausible human-like statements.
Evaluation of Predictive Reliability to Foster Trust in Artificial Intelligence. A case study in Multiple Sclerosis
Feb 27, 2024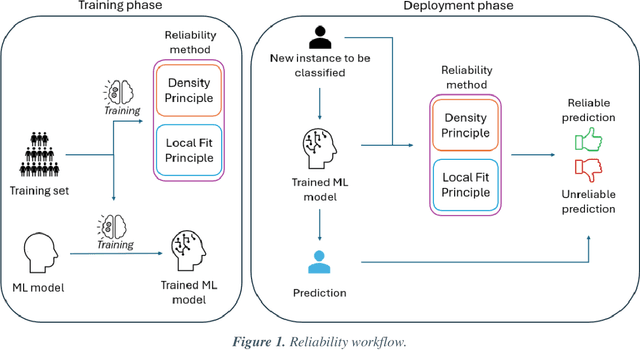
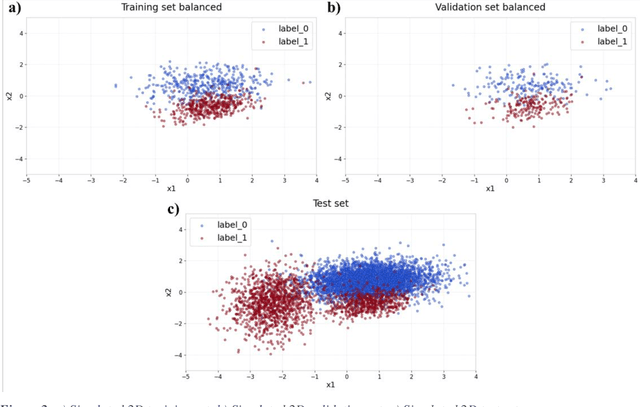
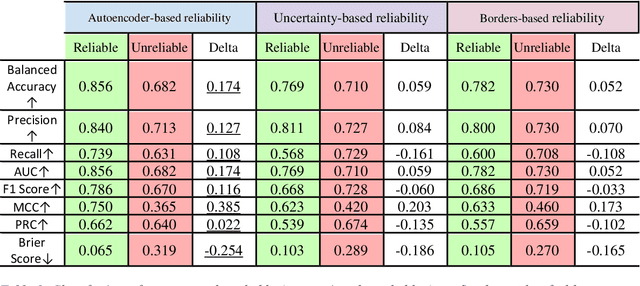
Abstract:Applying Artificial Intelligence (AI) and Machine Learning (ML) in critical contexts, such as medicine, requires the implementation of safety measures to reduce risks of harm in case of prediction errors. Spotting ML failures is of paramount importance when ML predictions are used to drive clinical decisions. ML predictive reliability measures the degree of trust of a ML prediction on a new instance, thus allowing decision-makers to accept or reject it based on its reliability. To assess reliability, we propose a method that implements two principles. First, our approach evaluates whether an instance to be classified is coming from the same distribution of the training set. To do this, we leverage Autoencoders (AEs) ability to reconstruct the training set with low error. An instance is considered Out-of-Distribution (OOD) if the AE reconstructs it with a high error. Second, it is evaluated whether the ML classifier has good performances on samples similar to the newly classified instance by using a proxy model. We show that this approach is able to assess reliability both in a simulated scenario and on a model trained to predict disease progression of Multiple Sclerosis patients. We also developed a Python package, named relAI, to embed reliability measures into ML pipelines. We propose a simple approach that can be used in the deployment phase of any ML model to suggest whether to trust predictions or not. Our method holds the promise to provide effective support to clinicians by spotting potential ML failures during deployment.
Advancing Italian Biomedical Information Extraction with Large Language Models: Methodological Insights and Multicenter Practical Application
Jun 08, 2023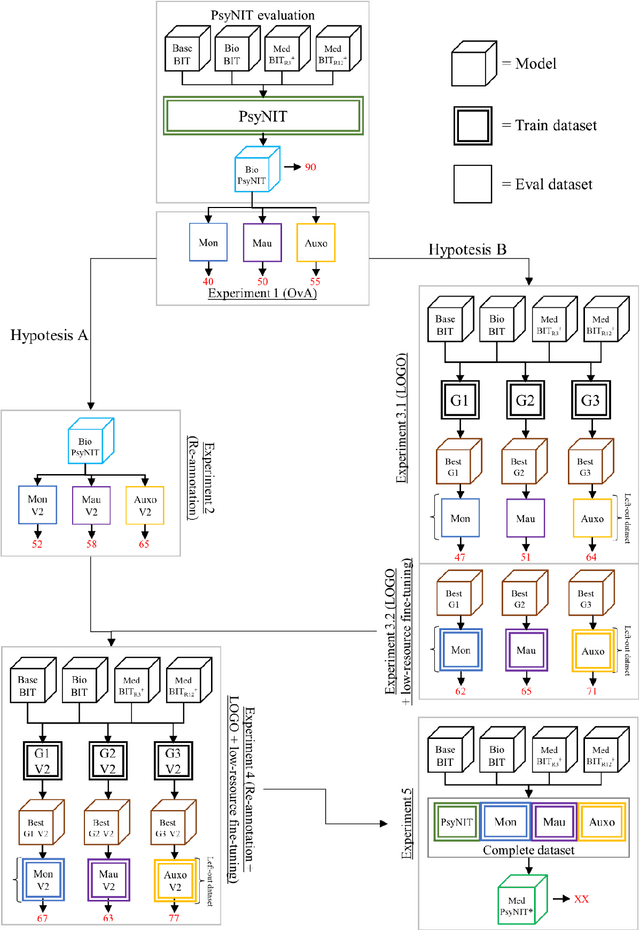


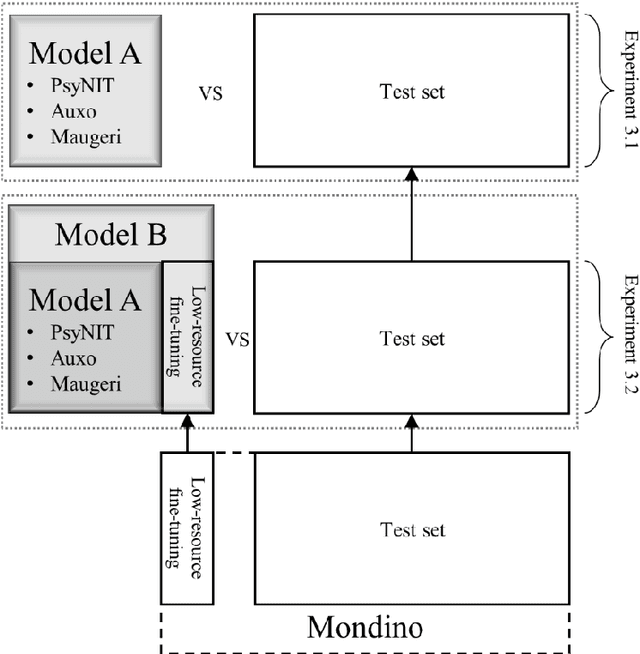
Abstract:The introduction of computerized medical records in hospitals has reduced burdensome operations like manual writing and information fetching. However, the data contained in medical records are still far underutilized, primarily because extracting them from unstructured textual medical records takes time and effort. Information Extraction, a subfield of Natural Language Processing, can help clinical practitioners overcome this limitation, using automated text-mining pipelines. In this work, we created the first Italian neuropsychiatric Named Entity Recognition dataset, PsyNIT, and used it to develop a Large Language Model for this task. Moreover, we conducted several experiments with three external independent datasets to implement an effective multicenter model, with overall F1-score 84.77%, Precision 83.16%, Recall 86.44%. The lessons learned are: (i) the crucial role of a consistent annotation process and (ii) a fine-tuning strategy that combines classical methods with a "few-shot" approach. This allowed us to establish methodological guidelines that pave the way for future implementations in this field and allow Italian hospitals to tap into important research opportunities.
Localising In-Domain Adaptation of Transformer-Based Biomedical Language Models
Dec 22, 2022Abstract:In the era of digital healthcare, the huge volumes of textual information generated every day in hospitals constitute an essential but underused asset that could be exploited with task-specific, fine-tuned biomedical language representation models, improving patient care and management. For such specialized domains, previous research has shown that fine-tuning models stemming from broad-coverage checkpoints can largely benefit additional training rounds over large-scale in-domain resources. However, these resources are often unreachable for less-resourced languages like Italian, preventing local medical institutions to employ in-domain adaptation. In order to reduce this gap, our work investigates two accessible approaches to derive biomedical language models in languages other than English, taking Italian as a concrete use-case: one based on neural machine translation of English resources, favoring quantity over quality; the other based on a high-grade, narrow-scoped corpus natively written in Italian, thus preferring quality over quantity. Our study shows that data quantity is a harder constraint than data quality for biomedical adaptation, but the concatenation of high-quality data can improve model performance even when dealing with relatively size-limited corpora. The models published from our investigations have the potential to unlock important research opportunities for Italian hospitals and academia. Finally, the set of lessons learned from the study constitutes valuable insights towards a solution to build biomedical language models that are generalizable to other less-resourced languages and different domain settings.
Tree-based local explanations of machine learning model predictions, AraucanaXAI
Oct 15, 2021Abstract:Increasingly complex learning methods such as boosting, bagging and deep learning have made ML models more accurate, but harder to understand and interpret. A tradeoff between performance and intelligibility is often to be faced, especially in high-stakes applications like medicine. In the present article we propose a novel methodological approach for generating explanations of the predictions of a generic ML model, given a specific instance for which the prediction has been made, that can tackle both classification and regression tasks. Advantages of the proposed XAI approach include improved fidelity to the original model, the ability to deal with non-linear decision boundaries, and native support to both classification and regression problems
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge