Amedeo Chiribiri
Deep learning motion correction of quantitative stress perfusion cardiovascular magnetic resonance
Oct 01, 2025Abstract:Background: Quantitative stress perfusion cardiovascular magnetic resonance (CMR) is a powerful tool for assessing myocardial ischemia. Motion correction is essential for accurate pixel-wise mapping but traditional registration-based methods are slow and sensitive to acquisition variability, limiting robustness and scalability. Methods: We developed an unsupervised deep learning-based motion correction pipeline that replaces iterative registration with efficient one-shot estimation. The method corrects motion in three steps and uses robust principal component analysis to reduce contrast-related effects. It aligns the perfusion series and auxiliary images (arterial input function and proton density-weighted series). Models were trained and validated on multivendor data from 201 patients, with 38 held out for testing. Performance was assessed via temporal alignment and quantitative perfusion values, compared to a previously published registration-based method. Results: The deep learning approach significantly improved temporal smoothness of time-intensity curves (p<0.001). Myocardial alignment (Dice = 0.92 (0.04) and 0.91 (0.05)) was comparable to the baseline and superior to before registration (Dice = 0.80 (0.09), p<0.001). Perfusion maps showed reduced motion, with lower standard deviation in the myocardium (0.52 (0.39) ml/min/g) compared to baseline (0.55 (0.44) ml/min/g). Processing time was reduced 15-fold. Conclusion: This deep learning pipeline enables fast, robust motion correction for stress perfusion CMR, improving accuracy across dynamic and auxiliary images. Trained on multivendor data, it generalizes across sequences and may facilitate broader clinical adoption of quantitative perfusion imaging.
DCRA-Net: Attention-Enabled Reconstruction Model for Dynamic Fetal Cardiac MRI
Dec 19, 2024



Abstract:Dynamic fetal heart magnetic resonance imaging (MRI) presents unique challenges due to the fast heart rate of the fetus compared to adult subjects and uncontrolled fetal motion. This requires high temporal and spatial resolutions over a large field of view, in order to encompass surrounding maternal anatomy. In this work, we introduce Dynamic Cardiac Reconstruction Attention Network (DCRA-Net) - a novel deep learning model that employs attention mechanisms in spatial and temporal domains and temporal frequency representation of data to reconstruct the dynamics of the fetal heart from highly accelerated free-running (non-gated) MRI acquisitions. DCRA-Net was trained on retrospectively undersampled complex-valued cardiac MRIs from 42 fetal subjects and separately from 153 adult subjects, and evaluated on data from 14 fetal and 39 adult subjects respectively. Its performance was compared to L+S and k-GIN methods in both fetal and adult cases for an undersampling factor of 8x. The proposed network performed better than the comparators for both fetal and adult data, for both regular lattice and centrally weighted random undersampling. Aliased signals due to the undersampling were comprehensively resolved, and both the spatial details of the heart and its temporal dynamics were recovered with high fidelity. The highest performance was achieved when using lattice undersampling, data consistency and temporal frequency representation, yielding PSNR of 38 for fetal and 35 for adult cases. Our method is publicly available at https://github.com/denproc/DCRA-Net.
Improved 3D Whole Heart Geometry from Sparse CMR Slices
Aug 14, 2024Abstract:Cardiac magnetic resonance (CMR) imaging and computed tomography (CT) are two common non-invasive imaging methods for assessing patients with cardiovascular disease. CMR typically acquires multiple sparse 2D slices, with unavoidable respiratory motion artefacts between slices, whereas CT acquires isotropic dense data but uses ionising radiation. In this study, we explore the combination of Slice Shifting Algorithm (SSA), Spatial Transformer Network (STN), and Label Transformer Network (LTN) to: 1) correct respiratory motion between segmented slices, and 2) transform sparse segmentation data into dense segmentation. All combinations were validated using synthetic motion-corrupted CMR slice segmentation generated from CT in 1699 cases, where the dense CT serves as the ground truth. In 199 testing cases, SSA-LTN achieved the best results for Dice score and Huasdorff distance (94.0% and 4.7 mm respectively, average over 5 labels) but gave topological errors in 8 cases. STN was effective as a plug-in tool for correcting all topological errors with minimal impact on overall performance (93.5% and 5.0 mm respectively). SSA also proves to be a valuable plug-in tool, enhancing performance over both STN-based and LTN-based models. The code for these different combinations is available at https://github.com/XESchong/STACOM2024.
High-Resolution Maps of Left Atrial Displacements and Strains Estimated with 3D CINE MRI and Unsupervised Neural Networks
Dec 14, 2023



Abstract:The functional analysis of the left atrium (LA) is important for evaluating cardiac health and understanding diseases like atrial fibrillation. Cine MRI is ideally placed for the detailed 3D characterisation of LA motion and deformation, but it is lacking appropriate acquisition and analysis tools. In this paper, we present Analysis for Left Atrial Displacements and Deformations using unsupervIsed neural Networks, \textit{Aladdin}, to automatically and reliably characterise regional LA deformations from high-resolution 3D Cine MRI. The tool includes: an online few-shot segmentation network (Aladdin-S), an online unsupervised image registration network (Aladdin-R), and a strain calculations pipeline tailored to the LA. We create maps of LA Displacement Vector Field (DVF) magnitude and LA principal strain values from images of 10 healthy volunteers and 8 patients with cardiovascular disease (CVD). We additionally create an atlas of these biomarkers using the data from the healthy volunteers. Aladdin is able to accurately track the LA wall across the cardiac cycle and characterize its motion and deformation. The overall DVF magnitude and principal strain values are significantly higher in the healthy group vs CVD patients: $2.85 \pm 1.59~mm$ and $0.09 \pm 0.05$ vs $1.96 \pm 0.74~mm$ and $0.03 \pm 0.04$, respectively. The time course of these metrics is also different in the two groups, with a more marked active contraction phase observed in the healthy cohort. Finally, utilizing the LA atlas allows us to identify regional deviations from the population distribution that may indicate focal tissue abnormalities. The proposed tool for the quantification of novel regional LA deformation biomarkers should have important clinical applications. The source code, anonymized images, generated maps and atlas are publicly available: https://github.com/cgalaz01/aladdin_cmr_la.
Physics-informed self-supervised deep learning reconstruction for accelerated first-pass perfusion cardiac MRI
Jan 05, 2023Abstract:First-pass perfusion cardiac magnetic resonance (FPP-CMR) is becoming an essential non-invasive imaging method for detecting deficits of myocardial blood flow, allowing the assessment of coronary heart disease. Nevertheless, acquisitions suffer from relatively low spatial resolution and limited heart coverage. Compressed sensing (CS) methods have been proposed to accelerate FPP-CMR and achieve higher spatial resolution. However, the long reconstruction times have limited the widespread clinical use of CS in FPP-CMR. Deep learning techniques based on supervised learning have emerged as alternatives for speeding up reconstructions. However, these approaches require fully sampled data for training, which is not possible to obtain, particularly high-resolution FPP-CMR images. Here, we propose a physics-informed self-supervised deep learning FPP-CMR reconstruction approach for accelerating FPP-CMR scans and hence facilitate high spatial resolution imaging. The proposed method provides high-quality FPP-CMR images from 10x undersampled data without using fully sampled reference data.
* Workshop on Machine Learning for Medical Image Reconstruction, MLMIR 2021, held in Conjunction with MICCAI 2021
CardiSort: a convolutional neural network for cross vendor automated sorting of cardiac MR images
Sep 17, 2021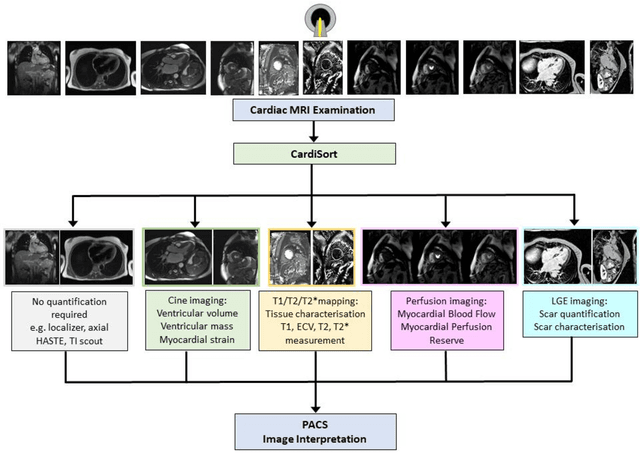
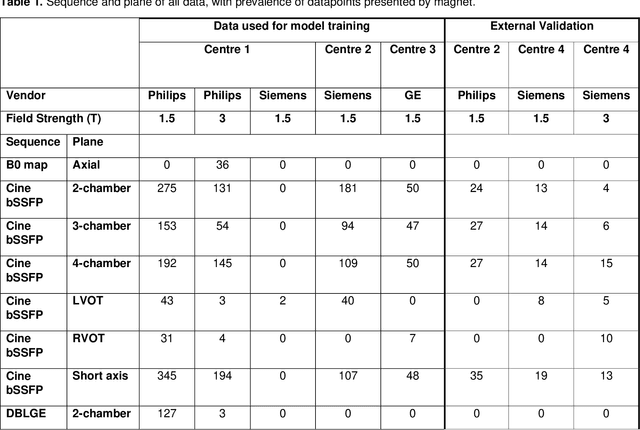
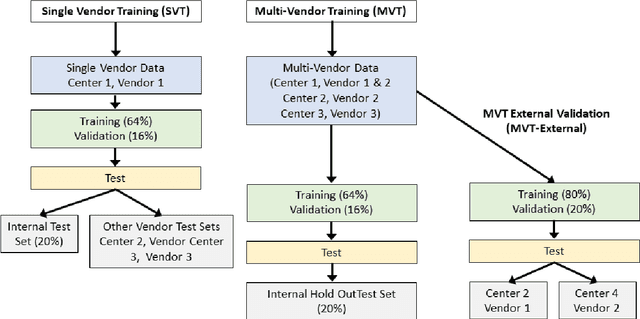
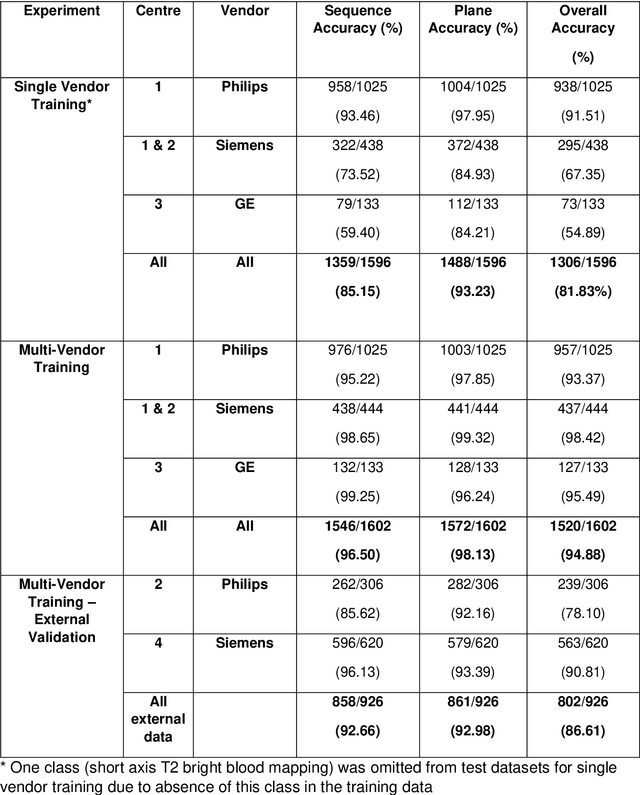
Abstract:Objectives: To develop an image-based automatic deep learning method to classify cardiac MR images by sequence type and imaging plane for improved clinical post-processing efficiency. Methods: Multi-vendor cardiac MRI studies were retrospectively collected from 4 centres and 3 vendors. A two-head convolutional neural network ('CardiSort') was trained to classify 35 sequences by imaging sequence (n=17) and plane (n=10). Single vendor training (SVT) on single centre images (n=234 patients) and multi-vendor training (MVT) with multicentre images (n = 479 patients, 3 centres) was performed. Model accuracy was compared to manual ground truth labels by an expert radiologist on a hold-out test set for both SVT and MVT. External validation of MVT (MVTexternal) was performed on data from 3 previously unseen magnet systems from 2 vendors (n=80 patients). Results: High sequence and plane accuracies were observed for SVT (85.2% and 93.2% respectively), and MVT (96.5% and 98.1% respectively) on the hold-out test set. MVTexternal yielded sequence accuracy of 92.7% and plane accuracy of 93.0%. There was high accuracy for common sequences and conventional cardiac planes. Poor accuracy was observed for underrepresented classes and sequences where there was greater variability in acquisition parameters across centres, such as perfusion imaging. Conclusions: A deep learning network was developed on multivendor data to classify MRI studies into component sequences and planes, with external validation. With refinement, it has potential to improve workflow by enabling automated sequence selection, an important first step in completely automated post-processing pipelines.
Physics-informed neural networks for myocardial perfusion MRI quantification
Dec 07, 2020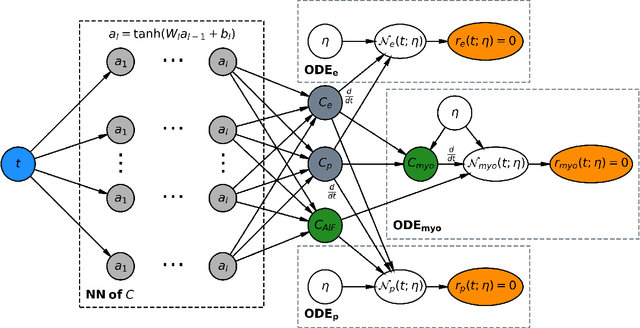
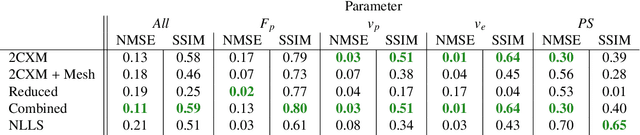
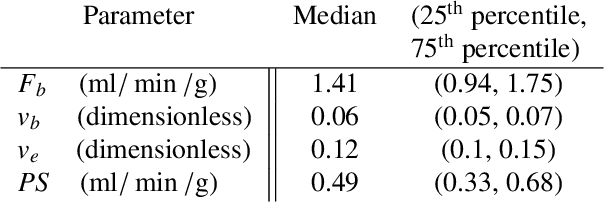
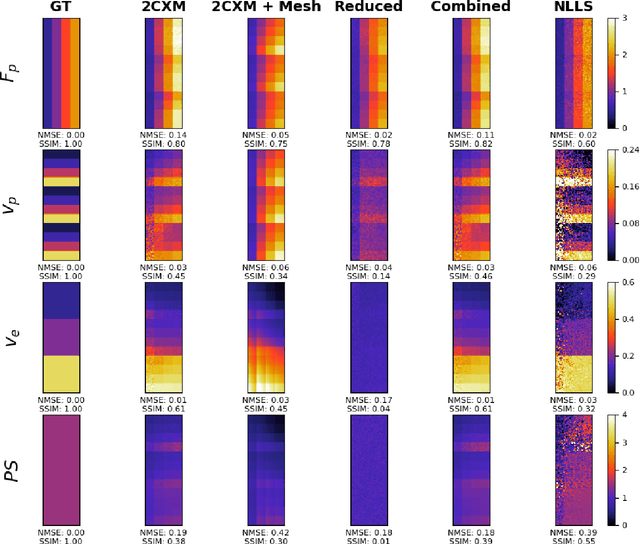
Abstract:Tracer-kinetic models allow for the quantification of kinetic parameters such as blood flow from dynamic contrast-enhanced magnetic resonance (MR) images. Fitting the observed data with multi-compartment exchange models is desirable, as they are physiologically plausible and resolve directly for blood flow and microvascular function. However, the reliability of model fitting is limited by the low signal-to-noise ratio, temporal resolution, and acquisition length. This may result in inaccurate parameter estimates. This study introduces physics-informed neural networks (PINNs) as a means to perform myocardial perfusion MR quantification, which provides a versatile scheme for the inference of kinetic parameters. These neural networks can be trained to fit the observed perfusion MR data while respecting the underlying physical conservation laws described by a multi-compartment exchange model. Here, we provide a framework for the implementation of PINNs in myocardial perfusion MR. The approach is validated both in silico and in vivo. In the in silico study, an overall reduction in mean-squared error with the ground-truth parameters was observed compared to a standard non-linear least squares fitting approach. The in vivo study demonstrates that the method produces parameter values comparable to those previously found in literature, as well as providing parameter maps which match the clinical diagnosis of patients.
Domain-Adversarial Learning for Multi-Centre, Multi-Vendor, and Multi-Disease Cardiac MR Image Segmentation
Aug 26, 2020
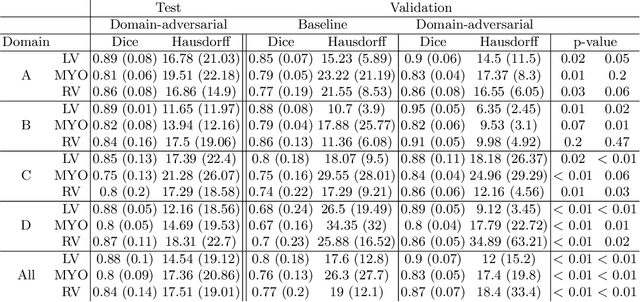
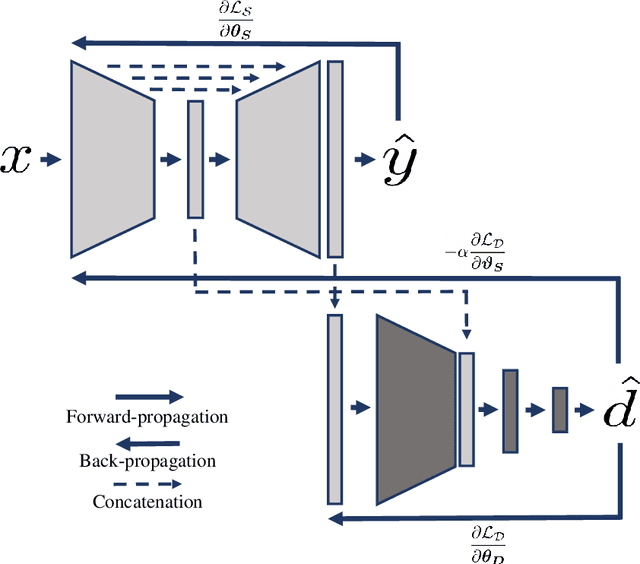
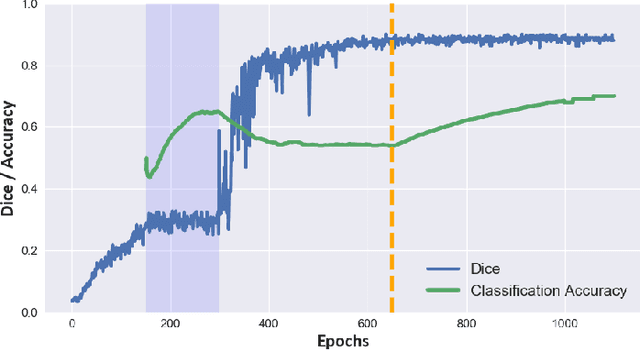
Abstract:Cine cardiac magnetic resonance (CMR) has become the gold standard for the non-invasive evaluation of cardiac function. In particular, it allows the accurate quantification of functional parameters including the chamber volumes and ejection fraction. Deep learning has shown the potential to automate the requisite cardiac structure segmentation. However, the lack of robustness of deep learning models has hindered their widespread clinical adoption. Due to differences in the data characteristics, neural networks trained on data from a specific scanner are not guaranteed to generalise well to data acquired at a different centre or with a different scanner. In this work, we propose a principled solution to the problem of this domain shift. Domain-adversarial learning is used to train a domain-invariant 2D U-Net using labelled and unlabelled data. This approach is evaluated on both seen and unseen domains from the M\&Ms challenge dataset and the domain-adversarial approach shows improved performance as compared to standard training. Additionally, we show that the domain information cannot be recovered from the learned features.
Deep learning-based prediction of kinetic parameters from myocardial perfusion MRI
Jul 27, 2019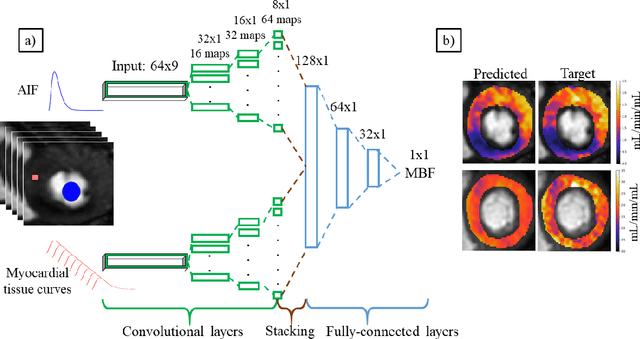
Abstract:The quantification of myocardial perfusion MRI has the potential to provide a fast, automated and user-independent assessment of myocardial ischaemia. However, due to the relatively high noise level and low temporal resolution of the acquired data and the complexity of the tracer-kinetic models, the model fitting can yield unreliable parameter estimates. A solution to this problem is the use of Bayesian inference which can incorporate prior knowledge and improve the reliability of the parameter estimation. This, however, uses Markov chain Monte Carlo sampling to approximate the posterior distribution of the kinetic parameters which is extremely time intensive. This work proposes training convolutional networks to directly predict the kinetic parameters from the signal-intensity curves that are trained using estimates obtained from the Bayesian inference. This allows fast estimation of the kinetic parameters with a similar performance to the Bayesian inference.
Hierarchical Bayesian myocardial perfusion quantification
Jun 06, 2019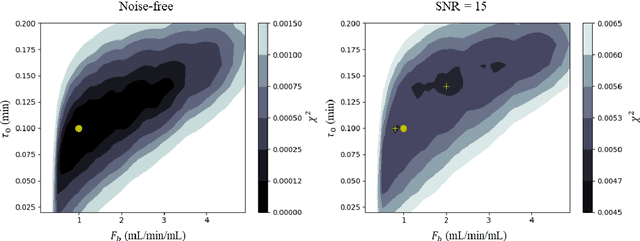
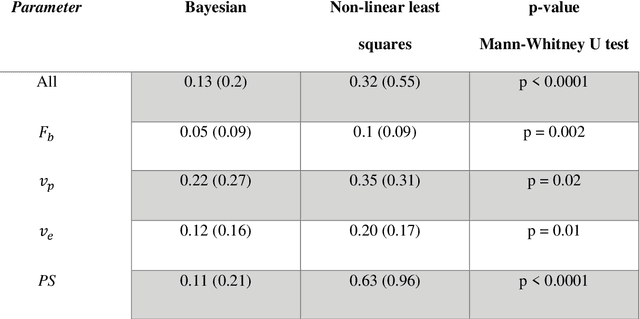
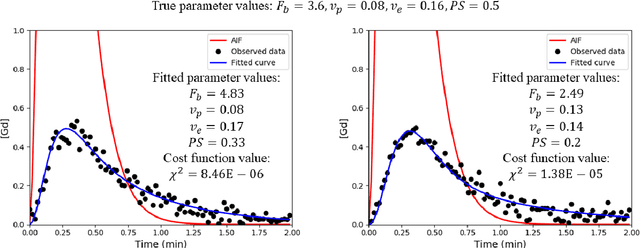
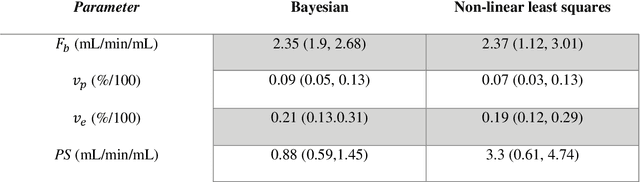
Abstract:Purpose: Tracer-kinetic models can be used for the quantitative assessment of contrast-enhanced MRI data. However, the model-fitting can produce unreliable results due to the limited data acquired and the high noise levels. Such problems are especially prevalent in myocardial perfusion MRI leading to the compromise of constrained numerical deconvolutions and segmental signal averaging being commonly used as alternatives to the more complex tracer-kinetic models. Methods: In this work, the use of hierarchical Bayesian inference for the parameter estimation is explored. It is shown that with Bayesian inference it is possible to reliably fit the two-compartment exchange model to perfusion data. The use of prior knowledge on the ranges of kinetic parameters and the fact that neighbouring voxels are likely to have similar kinetic properties combined with a Markov chain Monte Carlo based fitting procedure significantly improves the reliability of the perfusion estimates with compared to the traditional least-squares approach. The method is assessed using both simulated and patient data. Results: The average (standard deviation) normalised mean square error for the distinct noise realisations of a simulation phantom falls from 0.32 (0.55) with the least-squares fitting to 0.13 (0.2) using Bayesian inference. The assessment of the presence of coronary artery disease based purely on the quantitative MBF maps obtained using Bayesian inference matches the visual assessment in all 24 slices. When using the maps obtained by the least-squares fitting, a corresponding assessment is only achieved in 16/24 slices. Conclusion: Bayesian inference allows a reliable, fully automated and user-independent assessment of myocardial perfusion on a voxel-wise level using the two-compartment exchange model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge