Jack Lee
High-Resolution Maps of Left Atrial Displacements and Strains Estimated with 3D CINE MRI and Unsupervised Neural Networks
Dec 14, 2023



Abstract:The functional analysis of the left atrium (LA) is important for evaluating cardiac health and understanding diseases like atrial fibrillation. Cine MRI is ideally placed for the detailed 3D characterisation of LA motion and deformation, but it is lacking appropriate acquisition and analysis tools. In this paper, we present Analysis for Left Atrial Displacements and Deformations using unsupervIsed neural Networks, \textit{Aladdin}, to automatically and reliably characterise regional LA deformations from high-resolution 3D Cine MRI. The tool includes: an online few-shot segmentation network (Aladdin-S), an online unsupervised image registration network (Aladdin-R), and a strain calculations pipeline tailored to the LA. We create maps of LA Displacement Vector Field (DVF) magnitude and LA principal strain values from images of 10 healthy volunteers and 8 patients with cardiovascular disease (CVD). We additionally create an atlas of these biomarkers using the data from the healthy volunteers. Aladdin is able to accurately track the LA wall across the cardiac cycle and characterize its motion and deformation. The overall DVF magnitude and principal strain values are significantly higher in the healthy group vs CVD patients: $2.85 \pm 1.59~mm$ and $0.09 \pm 0.05$ vs $1.96 \pm 0.74~mm$ and $0.03 \pm 0.04$, respectively. The time course of these metrics is also different in the two groups, with a more marked active contraction phase observed in the healthy cohort. Finally, utilizing the LA atlas allows us to identify regional deviations from the population distribution that may indicate focal tissue abnormalities. The proposed tool for the quantification of novel regional LA deformation biomarkers should have important clinical applications. The source code, anonymized images, generated maps and atlas are publicly available: https://github.com/cgalaz01/aladdin_cmr_la.
Unsupervised Domain Adaptation for Automated Knee Osteoarthritis Phenotype Classification
Dec 14, 2022Abstract:Purpose: The aim of this study was to demonstrate the utility of unsupervised domain adaptation (UDA) in automated knee osteoarthritis (OA) phenotype classification using a small dataset (n=50). Materials and Methods: For this retrospective study, we collected 3,166 three-dimensional (3D) double-echo steady-state magnetic resonance (MR) images from the Osteoarthritis Initiative dataset and 50 3D turbo/fast spin-echo MR images from our institute (in 2020 and 2021) as the source and target datasets, respectively. For each patient, the degree of knee OA was initially graded according to the MRI Osteoarthritis Knee Score (MOAKS) before being converted to binary OA phenotype labels. The proposed UDA pipeline included (a) pre-processing, which involved automatic segmentation and region-of-interest cropping; (b) source classifier training, which involved pre-training phenotype classifiers on the source dataset; (c) target encoder adaptation, which involved unsupervised adaption of the source encoder to the target encoder and (d) target classifier validation, which involved statistical analysis of the target classification performance evaluated by the area under the receiver operating characteristic curve (AUROC), sensitivity, specificity and accuracy. Additionally, a classifier was trained without UDA for comparison. Results: The target classifier trained with UDA achieved improved AUROC, sensitivity, specificity and accuracy for both knee OA phenotypes compared with the classifier trained without UDA. Conclusion: The proposed UDA approach improves the performance of automated knee OA phenotype classification for small target datasets by utilising a large, high-quality source dataset for training. The results successfully demonstrated the advantages of the UDA approach in classification on small datasets.
Deep learning-based prediction of kinetic parameters from myocardial perfusion MRI
Jul 27, 2019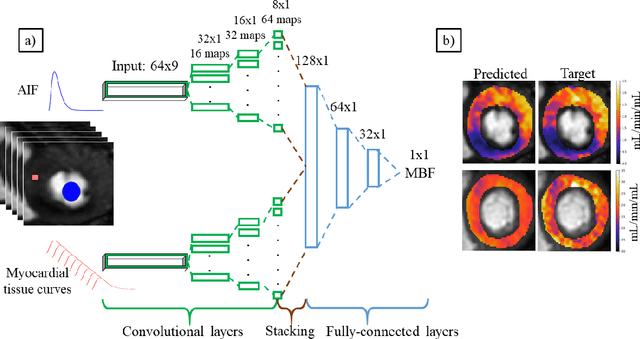
Abstract:The quantification of myocardial perfusion MRI has the potential to provide a fast, automated and user-independent assessment of myocardial ischaemia. However, due to the relatively high noise level and low temporal resolution of the acquired data and the complexity of the tracer-kinetic models, the model fitting can yield unreliable parameter estimates. A solution to this problem is the use of Bayesian inference which can incorporate prior knowledge and improve the reliability of the parameter estimation. This, however, uses Markov chain Monte Carlo sampling to approximate the posterior distribution of the kinetic parameters which is extremely time intensive. This work proposes training convolutional networks to directly predict the kinetic parameters from the signal-intensity curves that are trained using estimates obtained from the Bayesian inference. This allows fast estimation of the kinetic parameters with a similar performance to the Bayesian inference.
Hierarchical Bayesian myocardial perfusion quantification
Jun 06, 2019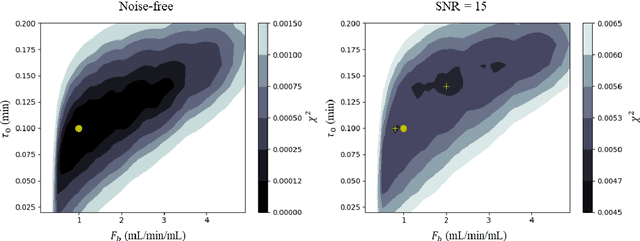
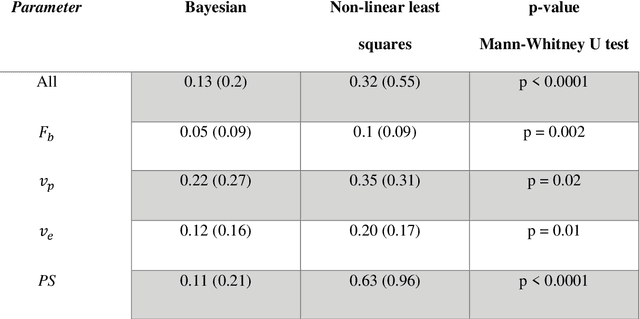
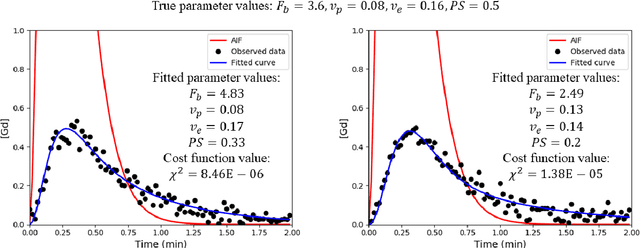
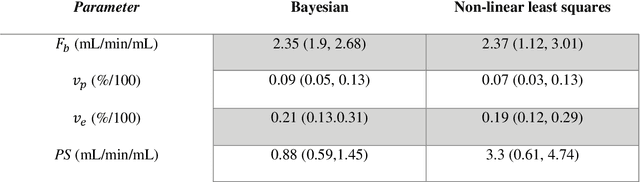
Abstract:Purpose: Tracer-kinetic models can be used for the quantitative assessment of contrast-enhanced MRI data. However, the model-fitting can produce unreliable results due to the limited data acquired and the high noise levels. Such problems are especially prevalent in myocardial perfusion MRI leading to the compromise of constrained numerical deconvolutions and segmental signal averaging being commonly used as alternatives to the more complex tracer-kinetic models. Methods: In this work, the use of hierarchical Bayesian inference for the parameter estimation is explored. It is shown that with Bayesian inference it is possible to reliably fit the two-compartment exchange model to perfusion data. The use of prior knowledge on the ranges of kinetic parameters and the fact that neighbouring voxels are likely to have similar kinetic properties combined with a Markov chain Monte Carlo based fitting procedure significantly improves the reliability of the perfusion estimates with compared to the traditional least-squares approach. The method is assessed using both simulated and patient data. Results: The average (standard deviation) normalised mean square error for the distinct noise realisations of a simulation phantom falls from 0.32 (0.55) with the least-squares fitting to 0.13 (0.2) using Bayesian inference. The assessment of the presence of coronary artery disease based purely on the quantitative MBF maps obtained using Bayesian inference matches the visual assessment in all 24 slices. When using the maps obtained by the least-squares fitting, a corresponding assessment is only achieved in 16/24 slices. Conclusion: Bayesian inference allows a reliable, fully automated and user-independent assessment of myocardial perfusion on a voxel-wise level using the two-compartment exchange model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge