Siyue Li
Department of Imaging and Interventional Radiology, the Chinese University of Hong Kong
MGML: A Plug-and-Play Meta-Guided Multi-Modal Learning Framework for Incomplete Multimodal Brain Tumor Segmentation
Dec 30, 2025Abstract:Leveraging multimodal information from Magnetic Resonance Imaging (MRI) plays a vital role in lesion segmentation, especially for brain tumors. However, in clinical practice, multimodal MRI data are often incomplete, making it challenging to fully utilize the available information. Therefore, maximizing the utilization of this incomplete multimodal information presents a crucial research challenge. We present a novel meta-guided multi-modal learning (MGML) framework that comprises two components: meta-parameterized adaptive modality fusion and consistency regularization module. The meta-parameterized adaptive modality fusion (Meta-AMF) enables the model to effectively integrate information from multiple modalities under varying input conditions. By generating adaptive soft-label supervision signals based on the available modalities, Meta-AMF explicitly promotes more coherent multimodal fusion. In addition, the consistency regularization module enhances segmentation performance and implicitly reinforces the robustness and generalization of the overall framework. Notably, our approach does not alter the original model architecture and can be conveniently integrated into the training pipeline for end-to-end model optimization. We conducted extensive experiments on the public BraTS2020 and BraTS2023 datasets. Compared to multiple state-of-the-art methods from previous years, our method achieved superior performance. On BraTS2020, for the average Dice scores across fifteen missing modality combinations, building upon the baseline, our method obtained scores of 87.55, 79.36, and 62.67 for the whole tumor (WT), the tumor core (TC), and the enhancing tumor (ET), respectively. We have made our source code publicly available at https://github.com/worldlikerr/MGML.
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Utilizing 3D Fast Spin Echo Anatomical Imaging to Reduce the Number of Contrast Preparations in $T_{1ρ}$ Quantification of Knee Cartilage Using Learning-Based Methods
Feb 13, 2025
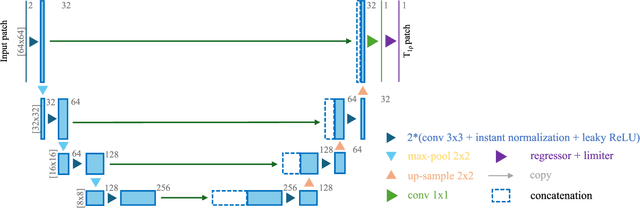
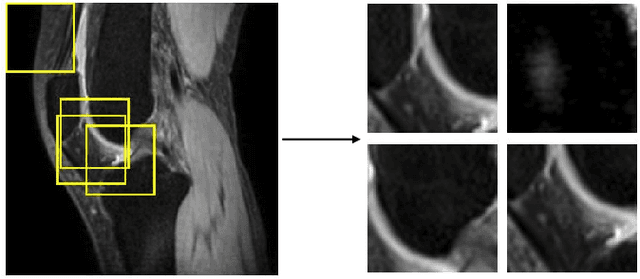
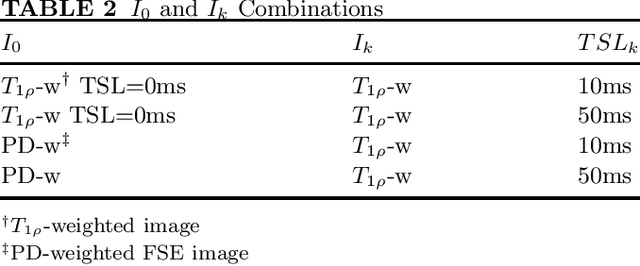
Abstract:Purpose: To propose and evaluate an accelerated $T_{1\rho}$ quantification method that combines $T_{1\rho}$-weighted fast spin echo (FSE) images and proton density (PD)-weighted anatomical FSE images, leveraging deep learning models for $T_{1\rho}$ mapping. The goal is to reduce scan time and facilitate integration into routine clinical workflows for osteoarthritis (OA) assessment. Methods: This retrospective study utilized MRI data from 40 participants (30 OA patients and 10 healthy volunteers). A volume of PD-weighted anatomical FSE images and a volume of $T_{1\rho}$-weighted images acquired at a non-zero spin-lock time were used as input to train deep learning models, including a 2D U-Net and a multi-layer perceptron (MLP). $T_{1\rho}$ maps generated by these models were compared with ground truth maps derived from a traditional non-linear least squares (NLLS) fitting method using four $T_{1\rho}$-weighted images. Evaluation metrics included mean absolute error (MAE), mean absolute percentage error (MAPE), regional error (RE), and regional percentage error (RPE). Results: Deep learning models achieved RPEs below 5% across all evaluated scenarios, outperforming NLLS methods, especially in low signal-to-noise conditions. The best results were obtained using the 2D U-Net, which effectively leveraged spatial information for accurate $T_{1\rho}$ fitting. The proposed method demonstrated compatibility with shorter TSLs, alleviating RF hardware and specific absorption rate (SAR) limitations. Conclusion: The proposed approach enables efficient $T_{1\rho}$ mapping using PD-weighted anatomical images, reducing scan time while maintaining clinical standards. This method has the potential to facilitate the integration of quantitative MRI techniques into routine clinical practice, benefiting OA diagnosis and monitoring.
ERANet: Edge Replacement Augmentation for Semi-Supervised Meniscus Segmentation with Prototype Consistency Alignment and Conditional Self-Training
Feb 11, 2025Abstract:Manual segmentation is labor-intensive, and automatic segmentation remains challenging due to the inherent variability in meniscal morphology, partial volume effects, and low contrast between the meniscus and surrounding tissues. To address these challenges, we propose ERANet, an innovative semi-supervised framework for meniscus segmentation that effectively leverages both labeled and unlabeled images through advanced augmentation and learning strategies. ERANet integrates three key components: edge replacement augmentation (ERA), prototype consistency alignment (PCA), and a conditional self-training (CST) strategy within a mean teacher architecture. ERA introduces anatomically relevant perturbations by simulating meniscal variations, ensuring that augmentations align with the structural context. PCA enhances segmentation performance by aligning intra-class features and promoting compact, discriminative feature representations, particularly in scenarios with limited labeled data. CST improves segmentation robustness by iteratively refining pseudo-labels and mitigating the impact of label noise during training. Together, these innovations establish ERANet as a robust and scalable solution for meniscus segmentation, effectively addressing key barriers to practical implementation. We validated ERANet comprehensively on 3D Double Echo Steady State (DESS) and 3D Fast/Turbo Spin Echo (FSE/TSE) MRI sequences. The results demonstrate the superior performance of ERANet compared to state-of-the-art methods. The proposed framework achieves reliable and accurate segmentation of meniscus structures, even when trained on minimal labeled data. Extensive ablation studies further highlight the synergistic contributions of ERA, PCA, and CST, solidifying ERANet as a transformative solution for semi-supervised meniscus segmentation in medical imaging.
DGSSA: Domain generalization with structural and stylistic augmentation for retinal vessel segmentation
Jan 07, 2025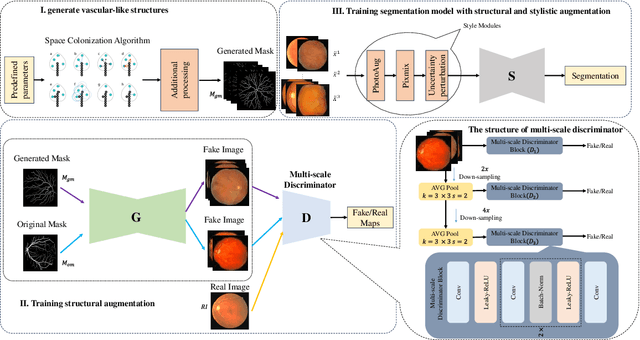
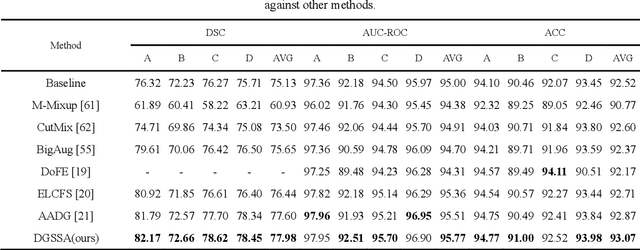
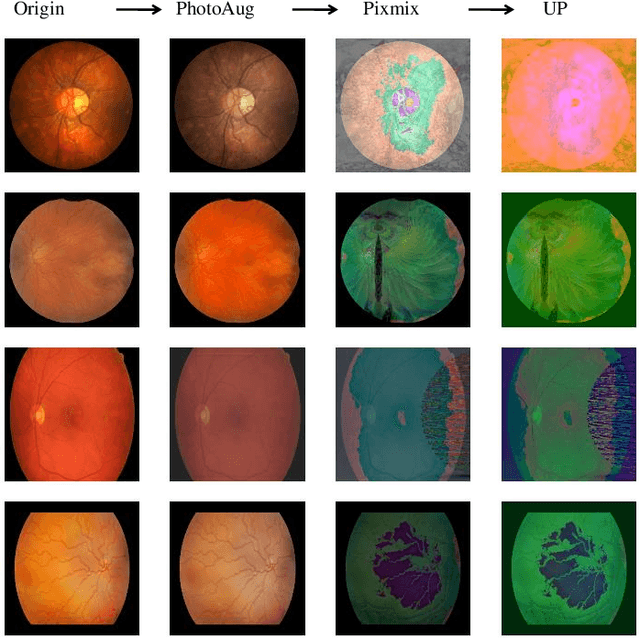
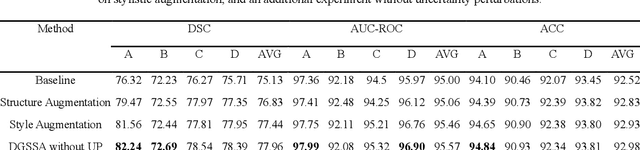
Abstract:Retinal vascular morphology is crucial for diagnosing diseases such as diabetes, glaucoma, and hypertension, making accurate segmentation of retinal vessels essential for early intervention. Traditional segmentation methods assume that training and testing data share similar distributions, which can lead to poor performance on unseen domains due to domain shifts caused by variations in imaging devices and patient demographics. This paper presents a novel approach, DGSSA, for retinal vessel image segmentation that enhances model generalization by combining structural and style augmentation strategies. We utilize a space colonization algorithm to generate diverse vascular-like structures that closely mimic actual retinal vessels, which are then used to generate pseudo-retinal images with an improved Pix2Pix model, allowing the segmentation model to learn a broader range of structure distributions. Additionally, we utilize PixMix to implement random photometric augmentations and introduce uncertainty perturbations, thereby enriching stylistic diversity and significantly enhancing the model's adaptability to varying imaging conditions. Our framework has been rigorously evaluated on four challenging datasets-DRIVE, CHASEDB, HRF, and STARE-demonstrating state-of-the-art performance that surpasses existing methods. This validates the effectiveness of our proposed approach, highlighting its potential for clinical application in automated retinal vessel analysis.
Unsupervised Domain Adaptation for Automated Knee Osteoarthritis Phenotype Classification
Dec 14, 2022Abstract:Purpose: The aim of this study was to demonstrate the utility of unsupervised domain adaptation (UDA) in automated knee osteoarthritis (OA) phenotype classification using a small dataset (n=50). Materials and Methods: For this retrospective study, we collected 3,166 three-dimensional (3D) double-echo steady-state magnetic resonance (MR) images from the Osteoarthritis Initiative dataset and 50 3D turbo/fast spin-echo MR images from our institute (in 2020 and 2021) as the source and target datasets, respectively. For each patient, the degree of knee OA was initially graded according to the MRI Osteoarthritis Knee Score (MOAKS) before being converted to binary OA phenotype labels. The proposed UDA pipeline included (a) pre-processing, which involved automatic segmentation and region-of-interest cropping; (b) source classifier training, which involved pre-training phenotype classifiers on the source dataset; (c) target encoder adaptation, which involved unsupervised adaption of the source encoder to the target encoder and (d) target classifier validation, which involved statistical analysis of the target classification performance evaluated by the area under the receiver operating characteristic curve (AUROC), sensitivity, specificity and accuracy. Additionally, a classifier was trained without UDA for comparison. Results: The target classifier trained with UDA achieved improved AUROC, sensitivity, specificity and accuracy for both knee OA phenotypes compared with the classifier trained without UDA. Conclusion: The proposed UDA approach improves the performance of automated knee OA phenotype classification for small target datasets by utilising a large, high-quality source dataset for training. The results successfully demonstrated the advantages of the UDA approach in classification on small datasets.
Denoising of Three-Dimensional Fast Spin Echo Magnetic Resonance Images of Knee Joints using Spatial-Variant Noise-Relevant Residual Learning of Convolution Neural Network
Apr 21, 2022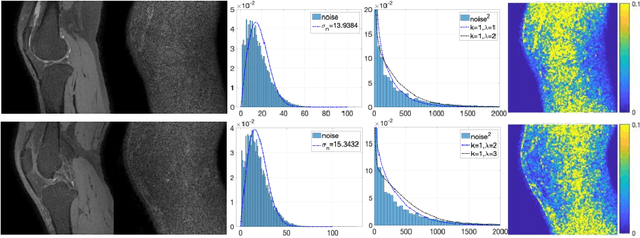
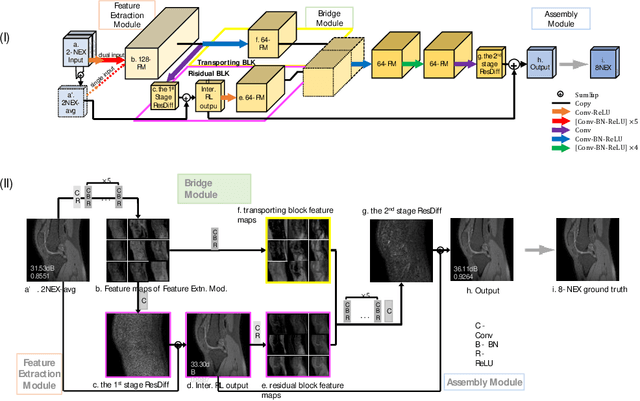
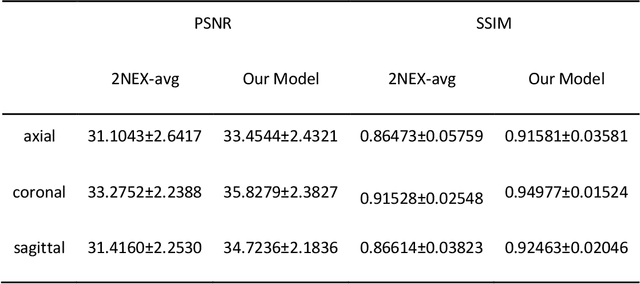

Abstract:Two-dimensional (2D) fast spin echo (FSE) techniques play a central role in the clinical magnetic resonance imaging (MRI) of knee joints. Moreover, three-dimensional (3D) FSE provides high-isotropic-resolution magnetic resonance (MR) images of knee joints, but it has a reduced signal-to-noise ratio compared to 2D FSE. Deep-learning denoising methods are a promising approach for denoising MR images, but they are often trained using synthetic noise due to challenges in obtaining true noise distributions for MR images. In this study, inherent true noise information from 2-NEX acquisition was used to develop a deep-learning model based on residual learning of convolutional neural network (CNN), and this model was used to suppress the noise in 3D FSE MR images of knee joints. The proposed CNN used two-step residual learning over parallel transporting and residual blocks and was designed to comprehensively learn real noise features from 2-NEX training data. The results of an ablation study validated the network design. The new method achieved improved denoising performance of 3D FSE knee MR images compared with current state-of-the-art methods, based on the peak signal-to-noise ratio and structural similarity index measure. The improved image quality after denoising using the new method was verified by radiological evaluation. A deep CNN using the inherent spatial-varying noise information in 2-NEX acquisitions was developed. This method showed promise for clinical MRI assessments of the knee, and has potential applications for the assessment of other anatomical structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge