Shuihua Wang
MGML: A Plug-and-Play Meta-Guided Multi-Modal Learning Framework for Incomplete Multimodal Brain Tumor Segmentation
Dec 30, 2025Abstract:Leveraging multimodal information from Magnetic Resonance Imaging (MRI) plays a vital role in lesion segmentation, especially for brain tumors. However, in clinical practice, multimodal MRI data are often incomplete, making it challenging to fully utilize the available information. Therefore, maximizing the utilization of this incomplete multimodal information presents a crucial research challenge. We present a novel meta-guided multi-modal learning (MGML) framework that comprises two components: meta-parameterized adaptive modality fusion and consistency regularization module. The meta-parameterized adaptive modality fusion (Meta-AMF) enables the model to effectively integrate information from multiple modalities under varying input conditions. By generating adaptive soft-label supervision signals based on the available modalities, Meta-AMF explicitly promotes more coherent multimodal fusion. In addition, the consistency regularization module enhances segmentation performance and implicitly reinforces the robustness and generalization of the overall framework. Notably, our approach does not alter the original model architecture and can be conveniently integrated into the training pipeline for end-to-end model optimization. We conducted extensive experiments on the public BraTS2020 and BraTS2023 datasets. Compared to multiple state-of-the-art methods from previous years, our method achieved superior performance. On BraTS2020, for the average Dice scores across fifteen missing modality combinations, building upon the baseline, our method obtained scores of 87.55, 79.36, and 62.67 for the whole tumor (WT), the tumor core (TC), and the enhancing tumor (ET), respectively. We have made our source code publicly available at https://github.com/worldlikerr/MGML.
ERANet: Edge Replacement Augmentation for Semi-Supervised Meniscus Segmentation with Prototype Consistency Alignment and Conditional Self-Training
Feb 11, 2025Abstract:Manual segmentation is labor-intensive, and automatic segmentation remains challenging due to the inherent variability in meniscal morphology, partial volume effects, and low contrast between the meniscus and surrounding tissues. To address these challenges, we propose ERANet, an innovative semi-supervised framework for meniscus segmentation that effectively leverages both labeled and unlabeled images through advanced augmentation and learning strategies. ERANet integrates three key components: edge replacement augmentation (ERA), prototype consistency alignment (PCA), and a conditional self-training (CST) strategy within a mean teacher architecture. ERA introduces anatomically relevant perturbations by simulating meniscal variations, ensuring that augmentations align with the structural context. PCA enhances segmentation performance by aligning intra-class features and promoting compact, discriminative feature representations, particularly in scenarios with limited labeled data. CST improves segmentation robustness by iteratively refining pseudo-labels and mitigating the impact of label noise during training. Together, these innovations establish ERANet as a robust and scalable solution for meniscus segmentation, effectively addressing key barriers to practical implementation. We validated ERANet comprehensively on 3D Double Echo Steady State (DESS) and 3D Fast/Turbo Spin Echo (FSE/TSE) MRI sequences. The results demonstrate the superior performance of ERANet compared to state-of-the-art methods. The proposed framework achieves reliable and accurate segmentation of meniscus structures, even when trained on minimal labeled data. Extensive ablation studies further highlight the synergistic contributions of ERA, PCA, and CST, solidifying ERANet as a transformative solution for semi-supervised meniscus segmentation in medical imaging.
WaveNet-SF: A Hybrid Network for Retinal Disease Detection Based on Wavelet Transform in the Spatial-Frequency Domain
Jan 21, 2025



Abstract:Retinal diseases are a leading cause of vision impairment and blindness, with timely diagnosis being critical for effective treatment. Optical Coherence Tomography (OCT) has become a standard imaging modality for retinal disease diagnosis, but OCT images often suffer from issues such as speckle noise, complex lesion shapes, and varying lesion sizes, making interpretation challenging. In this paper, we propose a novel framework, WaveNet-SF, to enhance retinal disease detection by integrating spatial-domain and frequency-domain learning. The framework utilizes wavelet transforms to decompose OCT images into low- and high-frequency components, enabling the model to extract both global structural features and fine-grained details. To improve lesion detection, we introduce a multi-scale wavelet spatial attention (MSW-SA) module, which enhances the model's focus on regions of interest at multiple scales. Additionally, a high-frequency feature compensation block (HFFC) is incorporated to recover edge information lost during wavelet decomposition, suppress noise, and preserve fine details crucial for lesion detection. Our approach achieves state-of-the-art (SOTA) classification accuracies of 97.82% and 99. 58% on the OCT-C8 and OCT2017 datasets, respectively, surpassing existing methods. These results demonstrate the efficacy of WaveNet-SF in addressing the challenges of OCT image analysis and its potential as a powerful tool for retinal disease diagnosis.
DGSSA: Domain generalization with structural and stylistic augmentation for retinal vessel segmentation
Jan 07, 2025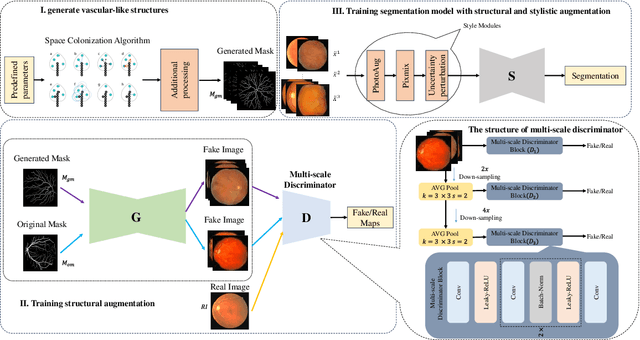
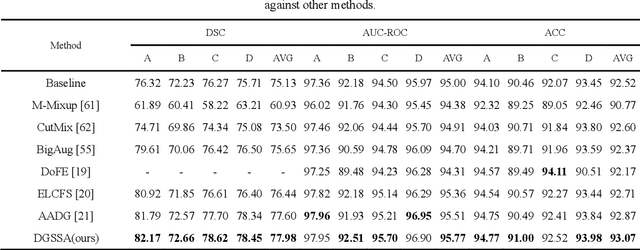
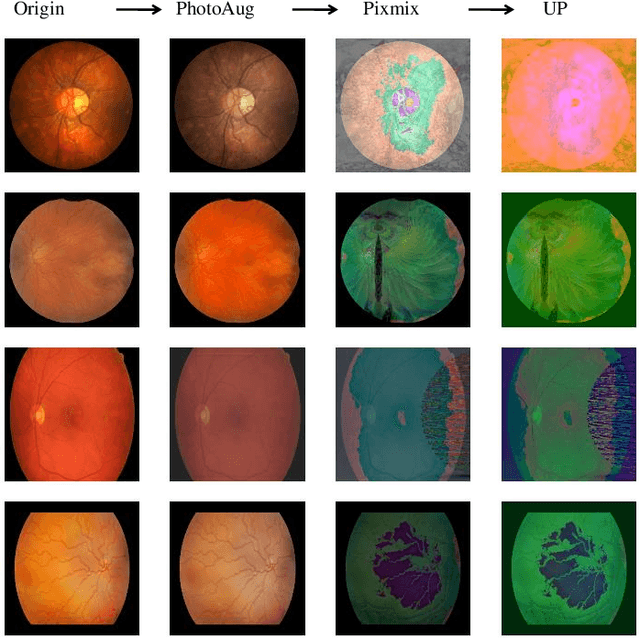
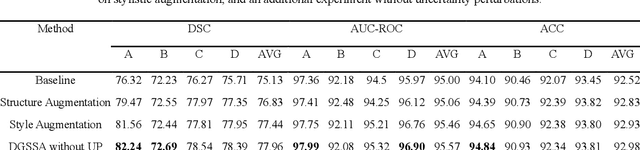
Abstract:Retinal vascular morphology is crucial for diagnosing diseases such as diabetes, glaucoma, and hypertension, making accurate segmentation of retinal vessels essential for early intervention. Traditional segmentation methods assume that training and testing data share similar distributions, which can lead to poor performance on unseen domains due to domain shifts caused by variations in imaging devices and patient demographics. This paper presents a novel approach, DGSSA, for retinal vessel image segmentation that enhances model generalization by combining structural and style augmentation strategies. We utilize a space colonization algorithm to generate diverse vascular-like structures that closely mimic actual retinal vessels, which are then used to generate pseudo-retinal images with an improved Pix2Pix model, allowing the segmentation model to learn a broader range of structure distributions. Additionally, we utilize PixMix to implement random photometric augmentations and introduce uncertainty perturbations, thereby enriching stylistic diversity and significantly enhancing the model's adaptability to varying imaging conditions. Our framework has been rigorously evaluated on four challenging datasets-DRIVE, CHASEDB, HRF, and STARE-demonstrating state-of-the-art performance that surpasses existing methods. This validates the effectiveness of our proposed approach, highlighting its potential for clinical application in automated retinal vessel analysis.
EAFP-Med: An Efficient Adaptive Feature Processing Module Based on Prompts for Medical Image Detection
Nov 27, 2023



Abstract:In the face of rapid advances in medical imaging, cross-domain adaptive medical image detection is challenging due to the differences in lesion representations across various medical imaging technologies. To address this issue, we draw inspiration from large language models to propose EAFP-Med, an efficient adaptive feature processing module based on prompts for medical image detection. EAFP-Med can efficiently extract lesion features of different scales from a diverse range of medical images based on prompts while being flexible and not limited by specific imaging techniques. Furthermore, it serves as a feature preprocessing module that can be connected to any model front-end to enhance the lesion features in input images. Moreover, we propose a novel adaptive disease detection model named EAFP-Med ST, which utilizes the Swin Transformer V2 - Tiny (SwinV2-T) as its backbone and connects it to EAFP-Med. We have compared our method to nine state-of-the-art methods. Experimental results demonstrate that EAFP-Med ST achieves the best performance on all three datasets (chest X-ray images, cranial magnetic resonance imaging images, and skin images). EAFP-Med can efficiently extract lesion features from various medical images based on prompts, enhancing the model's performance. This holds significant potential for improving medical image analysis and diagnosis.
Review of AlexNet for Medical Image Classification
Nov 15, 2023



Abstract:In recent years, the rapid development of deep learning has led to a wide range of applications in the field of medical image classification. The variants of neural network models with ever-increasing performance share some commonalities: to try to mitigate overfitting, improve generalization, avoid gradient vanishing and exploding, etc. AlexNet first utilizes the dropout technique to mitigate overfitting and the ReLU activation function to avoid gradient vanishing. Therefore, we focus our discussion on AlexNet, which has contributed greatly to the development of CNNs in 2012. After reviewing over 40 papers, including journal papers and conference papers, we give a narrative on the technical details, advantages, and application areas of AlexNet.
TBDLNet: a network for classifying multidrug-resistant and drug-sensitive tuberculosis
Oct 27, 2023



Abstract:This paper proposes applying a novel deep-learning model, TBDLNet, to recognize CT images to classify multidrug-resistant and drug-sensitive tuberculosis automatically. The pre-trained ResNet50 is selected to extract features. Three randomized neural networks are used to alleviate the overfitting problem. The ensemble of three RNNs is applied to boost the robustness via majority voting. The proposed model is evaluated by five-fold cross-validation. Five indexes are selected in this paper, which are accuracy, sensitivity, precision, F1-score, and specificity. The TBDLNet achieves 0.9822 accuracy, 0.9815 specificity, 0.9823 precision, 0.9829 sensitivity, and 0.9826 F1-score, respectively. The TBDLNet is suitable for classifying multidrug-resistant tuberculosis and drug-sensitive tuberculosis. It can detect multidrug-resistant pulmonary tuberculosis as early as possible, which helps to adjust the treatment plan in time and improve the treatment effect.
MEDNC: Multi-ensemble deep neural network for COVID-19 diagnosis
Apr 25, 2023



Abstract:Coronavirus disease 2019 (COVID-19) has spread all over the world for three years, but medical facilities in many areas still aren't adequate. There is a need for rapid COVID-19 diagnosis to identify high-risk patients and maximize the use of limited medical resources. Motivated by this fact, we proposed the deep learning framework MEDNC for automatic prediction and diagnosis of COVID-19 using computed tomography (CT) images. Our model was trained using two publicly available sets of COVID-19 data. And it was built with the inspiration of transfer learning. Results indicated that the MEDNC greatly enhanced the detection of COVID-19 infections, reaching an accuracy of 98.79% and 99.82% respectively. We tested MEDNC on a brain tumor and a blood cell dataset to show that our model applies to a wide range of problems. The outcomes demonstrated that our proposed models attained an accuracy of 99.39% and 99.28%, respectively. This COVID-19 recognition tool could help optimize healthcare resources and reduce clinicians' workload when screening for the virus.
MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images
Dec 28, 2022Abstract:A "heart attack" or myocardial infarction (MI), occurs when an artery supplying blood to the heart is abruptly occluded. The "gold standard" method for imaging MI is Cardiovascular Magnetic Resonance Imaging (MRI), with intravenously administered gadolinium-based contrast (late gadolinium enhancement). However, no "gold standard" fully automated method for the quantification of MI exists. In this work, we propose an end-to-end fully automatic system (MyI-Net) for the detection and quantification of MI in MRI images. This has the potential to reduce the uncertainty due to the technical variability across labs and inherent problems of the data and labels. Our system consists of four processing stages designed to maintain the flow of information across scales. First, features from raw MRI images are generated using feature extractors built on ResNet and MoblieNet architectures. This is followed by the Atrous Spatial Pyramid Pooling (ASPP) to produce spatial information at different scales to preserve more image context. High-level features from ASPP and initial low-level features are concatenated at the third stage and then passed to the fourth stage where spatial information is recovered via up-sampling to produce final image segmentation output into: i) background, ii) heart muscle, iii) blood and iv) scar areas. New models were compared with state-of-art models and manual quantification. Our models showed favorable performance in global segmentation and scar tissue detection relative to state-of-the-art work, including a four-fold better performance in matching scar pixels to contours produced by clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge