Yongcheng Yao
ERANet: Edge Replacement Augmentation for Semi-Supervised Meniscus Segmentation with Prototype Consistency Alignment and Conditional Self-Training
Feb 11, 2025Abstract:Manual segmentation is labor-intensive, and automatic segmentation remains challenging due to the inherent variability in meniscal morphology, partial volume effects, and low contrast between the meniscus and surrounding tissues. To address these challenges, we propose ERANet, an innovative semi-supervised framework for meniscus segmentation that effectively leverages both labeled and unlabeled images through advanced augmentation and learning strategies. ERANet integrates three key components: edge replacement augmentation (ERA), prototype consistency alignment (PCA), and a conditional self-training (CST) strategy within a mean teacher architecture. ERA introduces anatomically relevant perturbations by simulating meniscal variations, ensuring that augmentations align with the structural context. PCA enhances segmentation performance by aligning intra-class features and promoting compact, discriminative feature representations, particularly in scenarios with limited labeled data. CST improves segmentation robustness by iteratively refining pseudo-labels and mitigating the impact of label noise during training. Together, these innovations establish ERANet as a robust and scalable solution for meniscus segmentation, effectively addressing key barriers to practical implementation. We validated ERANet comprehensively on 3D Double Echo Steady State (DESS) and 3D Fast/Turbo Spin Echo (FSE/TSE) MRI sequences. The results demonstrate the superior performance of ERANet compared to state-of-the-art methods. The proposed framework achieves reliable and accurate segmentation of meniscus structures, even when trained on minimal labeled data. Extensive ablation studies further highlight the synergistic contributions of ERA, PCA, and CST, solidifying ERANet as a transformative solution for semi-supervised meniscus segmentation in medical imaging.
Quantifying Knee Cartilage Shape and Lesion: From Image to Metrics
Sep 11, 2024



Abstract:Imaging features of knee articular cartilage have been shown to be potential imaging biomarkers for knee osteoarthritis. Despite recent methodological advancements in image analysis techniques like image segmentation, registration, and domain-specific image computing algorithms, only a few works focus on building fully automated pipelines for imaging feature extraction. In this study, we developed a deep-learning-based medical image analysis application for knee cartilage morphometrics, CartiMorph Toolbox (CMT). We proposed a 2-stage joint template learning and registration network, CMT-reg. We trained the model using the OAI-ZIB dataset and assessed its performance in template-to-image registration. The CMT-reg demonstrated competitive results compared to other state-of-the-art models. We integrated the proposed model into an automated pipeline for the quantification of cartilage shape and lesion (full-thickness cartilage loss, specifically). The toolbox provides a comprehensive, user-friendly solution for medical image analysis and data visualization. The software and models are available at https://github.com/YongchengYAO/CMT-AMAI24paper .
CartiMorph: a framework for automated knee articular cartilage morphometrics
Aug 03, 2023Abstract:We introduce CartiMorph, a framework for automated knee articular cartilage morphometrics. It takes an image as input and generates quantitative metrics for cartilage subregions, including the percentage of full-thickness cartilage loss (FCL), mean thickness, surface area, and volume. CartiMorph leverages the power of deep learning models for hierarchical image feature representation. Deep learning models were trained and validated for tissue segmentation, template construction, and template-to-image registration. We established methods for surface-normal-based cartilage thickness mapping, FCL estimation, and rule-based cartilage parcellation. Our cartilage thickness map showed less error in thin and peripheral regions. We evaluated the effectiveness of the adopted segmentation model by comparing the quantitative metrics obtained from model segmentation and those from manual segmentation. The root-mean-squared deviation of the FCL measurements was less than 8%, and strong correlations were observed for the mean thickness (Pearson's correlation coefficient $\rho \in [0.82,0.97]$), surface area ($\rho \in [0.82,0.98]$) and volume ($\rho \in [0.89,0.98]$) measurements. We compared our FCL measurements with those from a previous study and found that our measurements deviated less from the ground truths. We observed superior performance of the proposed rule-based cartilage parcellation method compared with the atlas-based approach. CartiMorph has the potential to promote imaging biomarkers discovery for knee osteoarthritis.
Abnormal Functional Brain Network Connectivity Associated with Alzheimer's Disease
May 16, 2023



Abstract:The study's objective is to explore the distinctions in the functional brain network connectivity between Alzheimer's Disease (AD) patients and normal controls using Functional Magnetic Resonance Imaging (fMRI). The study included 590 individuals, with 175 having AD dementia and 415 age-, gender-, and handedness-matched normal controls. The connectivity of functional brain networks was measured using ROI-to-ROI and ROI-to-Voxel connectivity analyses. The findings reveal a general decrease in functional connectivity among the AD group in comparison to the normal control group. These results advance our comprehension of AD pathophysiology and could assist in identifying AD biomarkers.
Altered Topological Properties of Functional Brain Network Associated with Alzheimer's Disease
May 16, 2023Abstract:Functional Magnetic Resonance Imaging (fMRI) is commonly utilized to study human brain activity, including abnormal functional properties related to neurodegenerative diseases. This study aims to investigate the differences in the topological properties of functional brain networks between individuals with Alzheimer's Disease (AD) and normal controls. A total of 590 subjects, consisting of 175 with AD dementia and 415 age-, gender-, and handedness-matched controls, were included. The topological properties of the brain network were quantified using graph-theory-based analyses. The results indicate abnormal network integration and segregation in the AD group. These findings enhance our understanding of AD pathophysiology from a functional brain network structure perspective and may aid in identifying AD biomarkers. Supplementary data to aid in the validation of this research are available at https://github.com/YongchengYAO/AD-FunctionalBrainNetwork.
Unsupervised Domain Adaptation for Automated Knee Osteoarthritis Phenotype Classification
Dec 14, 2022Abstract:Purpose: The aim of this study was to demonstrate the utility of unsupervised domain adaptation (UDA) in automated knee osteoarthritis (OA) phenotype classification using a small dataset (n=50). Materials and Methods: For this retrospective study, we collected 3,166 three-dimensional (3D) double-echo steady-state magnetic resonance (MR) images from the Osteoarthritis Initiative dataset and 50 3D turbo/fast spin-echo MR images from our institute (in 2020 and 2021) as the source and target datasets, respectively. For each patient, the degree of knee OA was initially graded according to the MRI Osteoarthritis Knee Score (MOAKS) before being converted to binary OA phenotype labels. The proposed UDA pipeline included (a) pre-processing, which involved automatic segmentation and region-of-interest cropping; (b) source classifier training, which involved pre-training phenotype classifiers on the source dataset; (c) target encoder adaptation, which involved unsupervised adaption of the source encoder to the target encoder and (d) target classifier validation, which involved statistical analysis of the target classification performance evaluated by the area under the receiver operating characteristic curve (AUROC), sensitivity, specificity and accuracy. Additionally, a classifier was trained without UDA for comparison. Results: The target classifier trained with UDA achieved improved AUROC, sensitivity, specificity and accuracy for both knee OA phenotypes compared with the classifier trained without UDA. Conclusion: The proposed UDA approach improves the performance of automated knee OA phenotype classification for small target datasets by utilising a large, high-quality source dataset for training. The results successfully demonstrated the advantages of the UDA approach in classification on small datasets.
Multipath CNN with alpha matte inference for knee tissue segmentation from MRI
Sep 29, 2021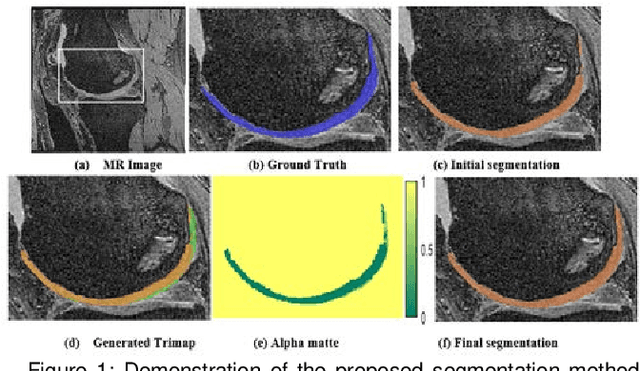
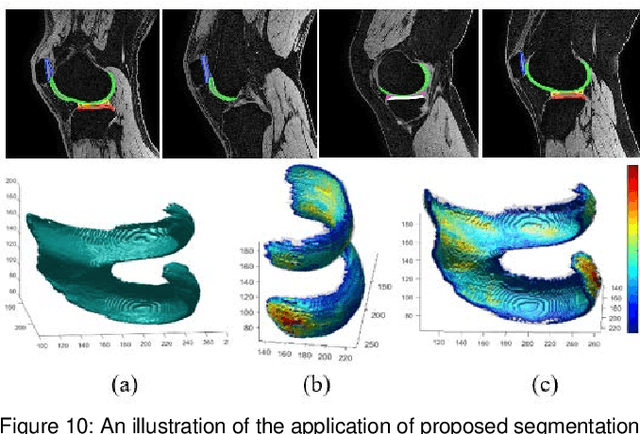
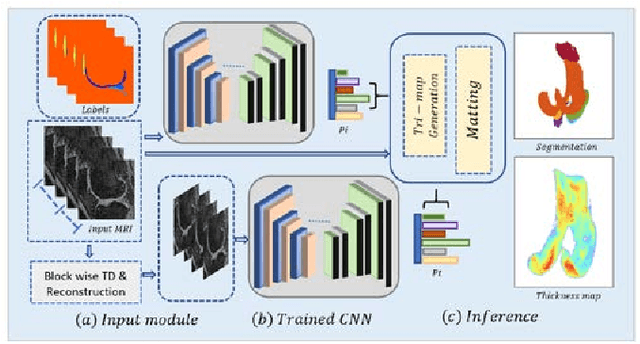
Abstract:Precise segmentation of knee tissues from magnetic resonance imaging (MRI) is critical in quantitative imaging and diagnosis. Convolutional neural networks (CNNs), which are state of the art, have limitations owing to the lack of image-specific adaptation, such as low tissue contrasts and structural inhomogeneities, thereby leading to incomplete segmentation results. This paper presents a deep learning based automatic segmentation framework for knee tissue segmentation. A novel multipath CNN-based method is proposed, which consists of an encoder decoder-based segmentation network in combination with a low rank tensor-reconstructed segmentation network. Low rank reconstruction in MRI tensor sub-blocks is introduced to exploit the structural and morphological variations in knee tissues. To further improve the segmentation from CNNs, trimap generation, which effectively utilizes superimposed regions, is proposed for defining high, medium and low confidence regions from the multipath CNNs. The secondary path with low rank reconstructed input mitigates the conditions in which the primary segmentation network can potentially fail and overlook the boundary regions. The outcome of the segmentation is solved as an alpha matting problem by blending the trimap with the source input. Experiments on Osteoarthritis Initiative (OAI) datasets and a self prepared scan validate the effectiveness of the proposed method. We specifically demonstrate the application of the proposed method in a cartilage segmentation based thickness map for diagnosis purposes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge