James F. Griffith
Utilizing 3D Fast Spin Echo Anatomical Imaging to Reduce the Number of Contrast Preparations in $T_{1ρ}$ Quantification of Knee Cartilage Using Learning-Based Methods
Feb 13, 2025
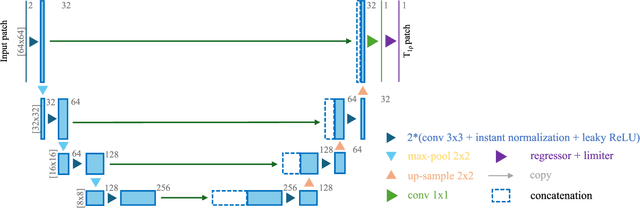
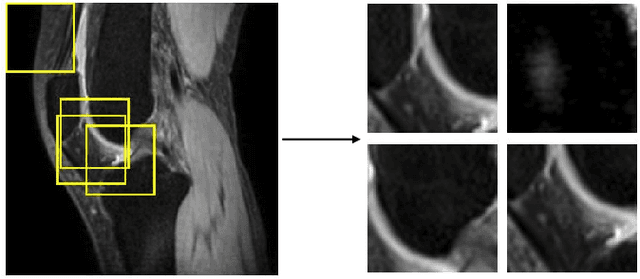
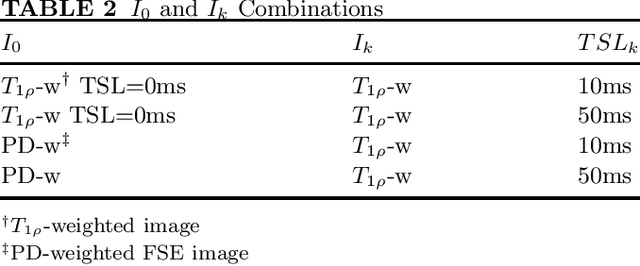
Abstract:Purpose: To propose and evaluate an accelerated $T_{1\rho}$ quantification method that combines $T_{1\rho}$-weighted fast spin echo (FSE) images and proton density (PD)-weighted anatomical FSE images, leveraging deep learning models for $T_{1\rho}$ mapping. The goal is to reduce scan time and facilitate integration into routine clinical workflows for osteoarthritis (OA) assessment. Methods: This retrospective study utilized MRI data from 40 participants (30 OA patients and 10 healthy volunteers). A volume of PD-weighted anatomical FSE images and a volume of $T_{1\rho}$-weighted images acquired at a non-zero spin-lock time were used as input to train deep learning models, including a 2D U-Net and a multi-layer perceptron (MLP). $T_{1\rho}$ maps generated by these models were compared with ground truth maps derived from a traditional non-linear least squares (NLLS) fitting method using four $T_{1\rho}$-weighted images. Evaluation metrics included mean absolute error (MAE), mean absolute percentage error (MAPE), regional error (RE), and regional percentage error (RPE). Results: Deep learning models achieved RPEs below 5% across all evaluated scenarios, outperforming NLLS methods, especially in low signal-to-noise conditions. The best results were obtained using the 2D U-Net, which effectively leveraged spatial information for accurate $T_{1\rho}$ fitting. The proposed method demonstrated compatibility with shorter TSLs, alleviating RF hardware and specific absorption rate (SAR) limitations. Conclusion: The proposed approach enables efficient $T_{1\rho}$ mapping using PD-weighted anatomical images, reducing scan time while maintaining clinical standards. This method has the potential to facilitate the integration of quantitative MRI techniques into routine clinical practice, benefiting OA diagnosis and monitoring.
Unsupervised Domain Adaptation for Automated Knee Osteoarthritis Phenotype Classification
Dec 14, 2022Abstract:Purpose: The aim of this study was to demonstrate the utility of unsupervised domain adaptation (UDA) in automated knee osteoarthritis (OA) phenotype classification using a small dataset (n=50). Materials and Methods: For this retrospective study, we collected 3,166 three-dimensional (3D) double-echo steady-state magnetic resonance (MR) images from the Osteoarthritis Initiative dataset and 50 3D turbo/fast spin-echo MR images from our institute (in 2020 and 2021) as the source and target datasets, respectively. For each patient, the degree of knee OA was initially graded according to the MRI Osteoarthritis Knee Score (MOAKS) before being converted to binary OA phenotype labels. The proposed UDA pipeline included (a) pre-processing, which involved automatic segmentation and region-of-interest cropping; (b) source classifier training, which involved pre-training phenotype classifiers on the source dataset; (c) target encoder adaptation, which involved unsupervised adaption of the source encoder to the target encoder and (d) target classifier validation, which involved statistical analysis of the target classification performance evaluated by the area under the receiver operating characteristic curve (AUROC), sensitivity, specificity and accuracy. Additionally, a classifier was trained without UDA for comparison. Results: The target classifier trained with UDA achieved improved AUROC, sensitivity, specificity and accuracy for both knee OA phenotypes compared with the classifier trained without UDA. Conclusion: The proposed UDA approach improves the performance of automated knee OA phenotype classification for small target datasets by utilising a large, high-quality source dataset for training. The results successfully demonstrated the advantages of the UDA approach in classification on small datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge