Basim Azam
Leveraging Text-to-Image Generation for Handling Spurious Correlation
Mar 21, 2025Abstract:Deep neural networks trained with Empirical Risk Minimization (ERM) perform well when both training and test data come from the same domain, but they often fail to generalize to out-of-distribution samples. In image classification, these models may rely on spurious correlations that often exist between labels and irrelevant features of images, making predictions unreliable when those features do not exist. We propose a technique to generate training samples with text-to-image (T2I) diffusion models for addressing the spurious correlation problem. First, we compute the best describing token for the visual features pertaining to the causal components of samples by a textual inversion mechanism. Then, leveraging a language segmentation method and a diffusion model, we generate new samples by combining the causal component with the elements from other classes. We also meticulously prune the generated samples based on the prediction probabilities and attribution scores of the ERM model to ensure their correct composition for our objective. Finally, we retrain the ERM model on our augmented dataset. This process reduces the model's reliance on spurious correlations by learning from carefully crafted samples for in which this correlation does not exist. Our experiments show that across different benchmarks, our technique achieves better worst-group accuracy than the existing state-of-the-art methods.
GO-N3RDet: Geometry Optimized NeRF-enhanced 3D Object Detector
Mar 19, 2025Abstract:We propose GO-N3RDet, a scene-geometry optimized multi-view 3D object detector enhanced by neural radiance fields. The key to accurate 3D object detection is in effective voxel representation. However, due to occlusion and lack of 3D information, constructing 3D features from multi-view 2D images is challenging. Addressing that, we introduce a unique 3D positional information embedded voxel optimization mechanism to fuse multi-view features. To prioritize neural field reconstruction in object regions, we also devise a double importance sampling scheme for the NeRF branch of our detector. We additionally propose an opacity optimization module for precise voxel opacity prediction by enforcing multi-view consistency constraints. Moreover, to further improve voxel density consistency across multiple perspectives, we incorporate ray distance as a weighting factor to minimize cumulative ray errors. Our unique modules synergetically form an end-to-end neural model that establishes new state-of-the-art in NeRF-based multi-view 3D detection, verified with extensive experiments on ScanNet and ARKITScenes. Code will be available at https://github.com/ZechuanLi/GO-N3RDet.
Suitability of KANs for Computer Vision: A preliminary investigation
Jun 13, 2024Abstract:Kolmogorov-Arnold Networks (KANs) introduce a paradigm of neural modeling that implements learnable functions on the edges of the networks, diverging from the traditional node-centric activations in neural networks. This work assesses the applicability and efficacy of KANs in visual modeling, focusing on the image recognition task. We mainly analyze the performance and efficiency of different network architectures built using KAN concepts along with conventional building blocks of convolutional and linear layers, enabling a comparative analysis with the conventional models. Our findings are aimed at contributing to understanding the potential of KANs in computer vision, highlighting both their strengths and areas for further research. Our evaluation shows that whereas KAN-based architectures perform in-line with the original claims of KAN paper for performance and model-complexity in the case of simpler vision datasets like MNIST, the advantages seem to diminish even for slightly more complex datasets like CIFAR-10.
Context-based Deep Learning Architecture with Optimal Integration Layer for Image Parsing
Apr 13, 2022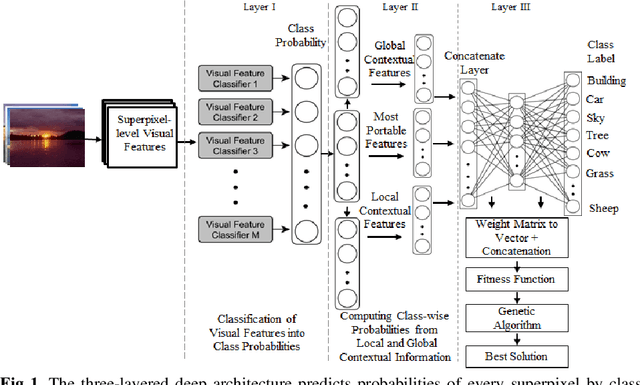


Abstract:Deep learning models have been efficient lately on image parsing tasks. However, deep learning models are not fully capable of exploiting visual and contextual information simultaneously. The proposed three-layer context-based deep architecture is capable of integrating context explicitly with visual information. The novel idea here is to have a visual layer to learn visual characteristics from binary class-based learners, a contextual layer to learn context, and then an integration layer to learn from both via genetic algorithm-based optimal fusion to produce a final decision. The experimental outcomes when evaluated on benchmark datasets are promising. Further analysis shows that optimized network weights can improve performance and make stable predictions.
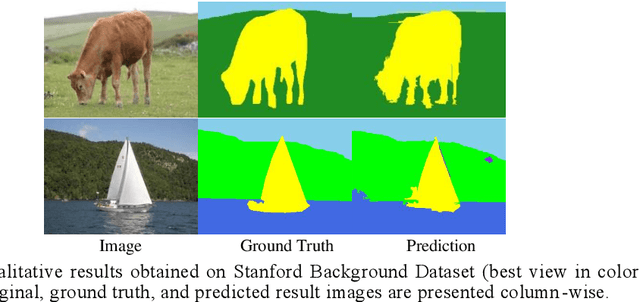
Deep Learning Model with GA based Feature Selection and Context Integration
Apr 13, 2022



Abstract:Deep learning models have been very successful in computer vision and image processing applications. Since its inception, Many top-performing methods for image segmentation are based on deep CNN models. However, deep CNN models fail to integrate global and local context alongside visual features despite having complex multi-layer architectures. We propose a novel three-layered deep learning model that assiminlate or learns independently global and local contextual information alongside visual features. The novelty of the proposed model is that One-vs-All binary class-based learners are introduced to learn Genetic Algorithm (GA) optimized features in the visual layer, followed by the contextual layer that learns global and local contexts of an image, and finally the third layer integrates all the information optimally to obtain the final class label. Stanford Background and CamVid benchmark image parsing datasets were used for our model evaluation, and our model shows promising results. The empirical analysis reveals that optimized visual features with global and local contextual information play a significant role to improve accuracy and produce stable predictions comparable to state-of-the-art deep CNN models.
Multipath CNN with alpha matte inference for knee tissue segmentation from MRI
Sep 29, 2021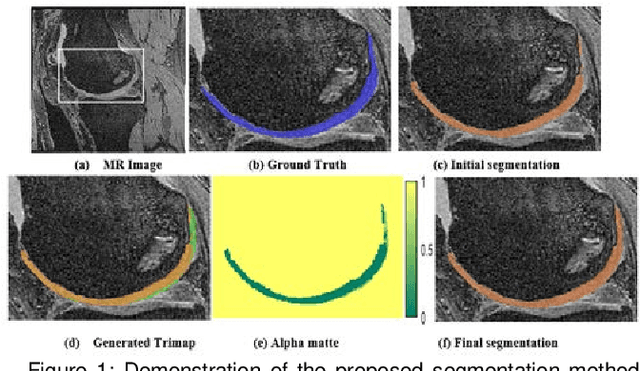
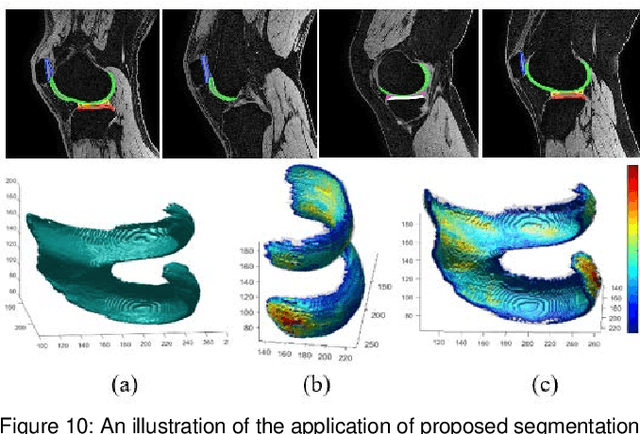
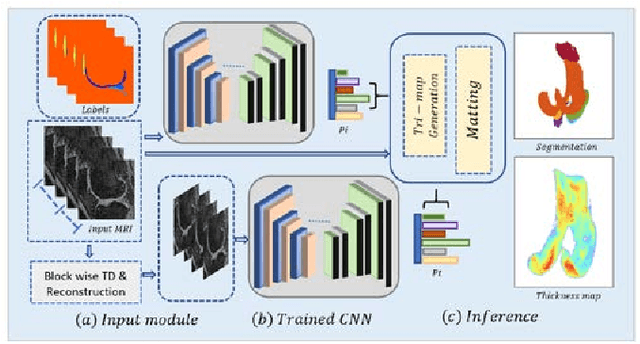
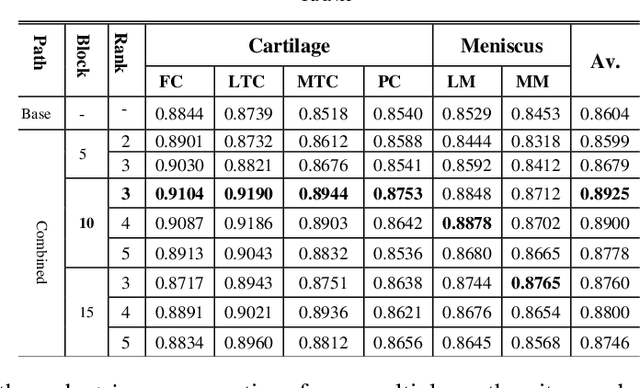
Abstract:Precise segmentation of knee tissues from magnetic resonance imaging (MRI) is critical in quantitative imaging and diagnosis. Convolutional neural networks (CNNs), which are state of the art, have limitations owing to the lack of image-specific adaptation, such as low tissue contrasts and structural inhomogeneities, thereby leading to incomplete segmentation results. This paper presents a deep learning based automatic segmentation framework for knee tissue segmentation. A novel multipath CNN-based method is proposed, which consists of an encoder decoder-based segmentation network in combination with a low rank tensor-reconstructed segmentation network. Low rank reconstruction in MRI tensor sub-blocks is introduced to exploit the structural and morphological variations in knee tissues. To further improve the segmentation from CNNs, trimap generation, which effectively utilizes superimposed regions, is proposed for defining high, medium and low confidence regions from the multipath CNNs. The secondary path with low rank reconstructed input mitigates the conditions in which the primary segmentation network can potentially fail and overlook the boundary regions. The outcome of the segmentation is solved as an alpha matting problem by blending the trimap with the source input. Experiments on Osteoarthritis Initiative (OAI) datasets and a self prepared scan validate the effectiveness of the proposed method. We specifically demonstrate the application of the proposed method in a cartilage segmentation based thickness map for diagnosis purposes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge