Shutian Zhao
Department of Imaging and Interventional Radiology, the Chinese University of Hong Kong
ERANet: Edge Replacement Augmentation for Semi-Supervised Meniscus Segmentation with Prototype Consistency Alignment and Conditional Self-Training
Feb 11, 2025Abstract:Manual segmentation is labor-intensive, and automatic segmentation remains challenging due to the inherent variability in meniscal morphology, partial volume effects, and low contrast between the meniscus and surrounding tissues. To address these challenges, we propose ERANet, an innovative semi-supervised framework for meniscus segmentation that effectively leverages both labeled and unlabeled images through advanced augmentation and learning strategies. ERANet integrates three key components: edge replacement augmentation (ERA), prototype consistency alignment (PCA), and a conditional self-training (CST) strategy within a mean teacher architecture. ERA introduces anatomically relevant perturbations by simulating meniscal variations, ensuring that augmentations align with the structural context. PCA enhances segmentation performance by aligning intra-class features and promoting compact, discriminative feature representations, particularly in scenarios with limited labeled data. CST improves segmentation robustness by iteratively refining pseudo-labels and mitigating the impact of label noise during training. Together, these innovations establish ERANet as a robust and scalable solution for meniscus segmentation, effectively addressing key barriers to practical implementation. We validated ERANet comprehensively on 3D Double Echo Steady State (DESS) and 3D Fast/Turbo Spin Echo (FSE/TSE) MRI sequences. The results demonstrate the superior performance of ERANet compared to state-of-the-art methods. The proposed framework achieves reliable and accurate segmentation of meniscus structures, even when trained on minimal labeled data. Extensive ablation studies further highlight the synergistic contributions of ERA, PCA, and CST, solidifying ERANet as a transformative solution for semi-supervised meniscus segmentation in medical imaging.
Denoising of Three-Dimensional Fast Spin Echo Magnetic Resonance Images of Knee Joints using Spatial-Variant Noise-Relevant Residual Learning of Convolution Neural Network
Apr 21, 2022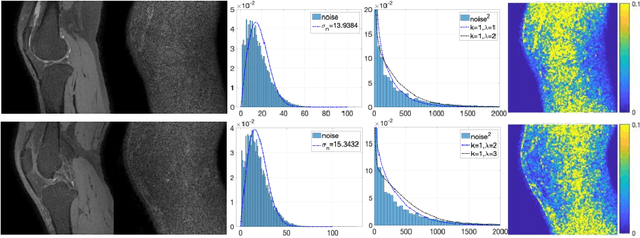
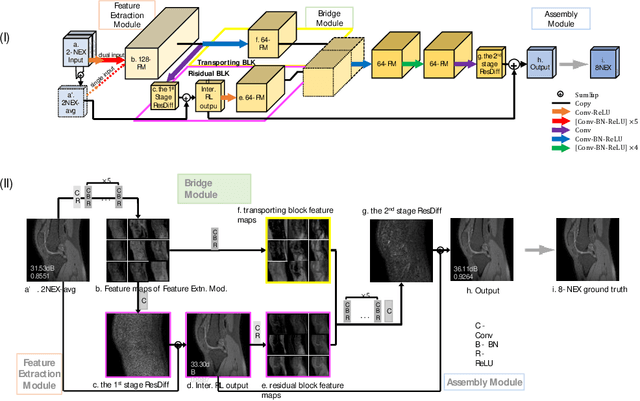
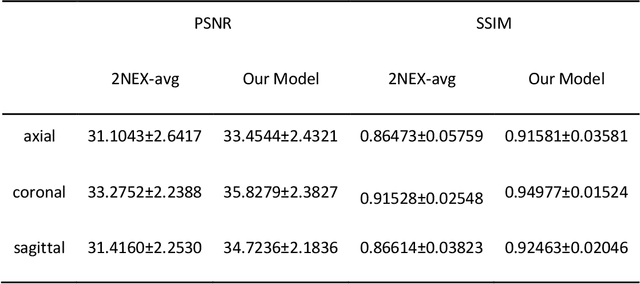

Abstract:Two-dimensional (2D) fast spin echo (FSE) techniques play a central role in the clinical magnetic resonance imaging (MRI) of knee joints. Moreover, three-dimensional (3D) FSE provides high-isotropic-resolution magnetic resonance (MR) images of knee joints, but it has a reduced signal-to-noise ratio compared to 2D FSE. Deep-learning denoising methods are a promising approach for denoising MR images, but they are often trained using synthetic noise due to challenges in obtaining true noise distributions for MR images. In this study, inherent true noise information from 2-NEX acquisition was used to develop a deep-learning model based on residual learning of convolutional neural network (CNN), and this model was used to suppress the noise in 3D FSE MR images of knee joints. The proposed CNN used two-step residual learning over parallel transporting and residual blocks and was designed to comprehensively learn real noise features from 2-NEX training data. The results of an ablation study validated the network design. The new method achieved improved denoising performance of 3D FSE knee MR images compared with current state-of-the-art methods, based on the peak signal-to-noise ratio and structural similarity index measure. The improved image quality after denoising using the new method was verified by radiological evaluation. A deep CNN using the inherent spatial-varying noise information in 2-NEX acquisitions was developed. This method showed promise for clinical MRI assessments of the knee, and has potential applications for the assessment of other anatomical structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge