Anil A. Bharath
Department of Bioengineering, Imperial College London
High-Resolution Maps of Left Atrial Displacements and Strains Estimated with 3D CINE MRI and Unsupervised Neural Networks
Dec 14, 2023



Abstract:The functional analysis of the left atrium (LA) is important for evaluating cardiac health and understanding diseases like atrial fibrillation. Cine MRI is ideally placed for the detailed 3D characterisation of LA motion and deformation, but it is lacking appropriate acquisition and analysis tools. In this paper, we present Analysis for Left Atrial Displacements and Deformations using unsupervIsed neural Networks, \textit{Aladdin}, to automatically and reliably characterise regional LA deformations from high-resolution 3D Cine MRI. The tool includes: an online few-shot segmentation network (Aladdin-S), an online unsupervised image registration network (Aladdin-R), and a strain calculations pipeline tailored to the LA. We create maps of LA Displacement Vector Field (DVF) magnitude and LA principal strain values from images of 10 healthy volunteers and 8 patients with cardiovascular disease (CVD). We additionally create an atlas of these biomarkers using the data from the healthy volunteers. Aladdin is able to accurately track the LA wall across the cardiac cycle and characterize its motion and deformation. The overall DVF magnitude and principal strain values are significantly higher in the healthy group vs CVD patients: $2.85 \pm 1.59~mm$ and $0.09 \pm 0.05$ vs $1.96 \pm 0.74~mm$ and $0.03 \pm 0.04$, respectively. The time course of these metrics is also different in the two groups, with a more marked active contraction phase observed in the healthy cohort. Finally, utilizing the LA atlas allows us to identify regional deviations from the population distribution that may indicate focal tissue abnormalities. The proposed tool for the quantification of novel regional LA deformation biomarkers should have important clinical applications. The source code, anonymized images, generated maps and atlas are publicly available: https://github.com/cgalaz01/aladdin_cmr_la.
TMS-Net: A Segmentation Network Coupled With A Run-time Quality Control Method For Robust Cardiac Image Segmentation
Dec 21, 2022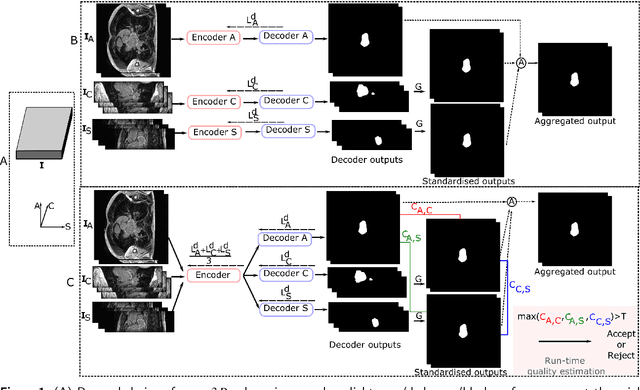

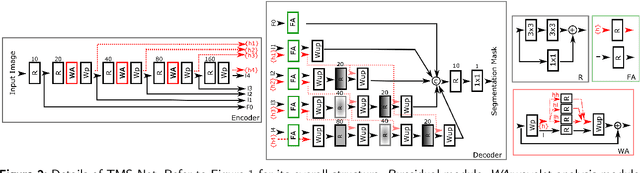
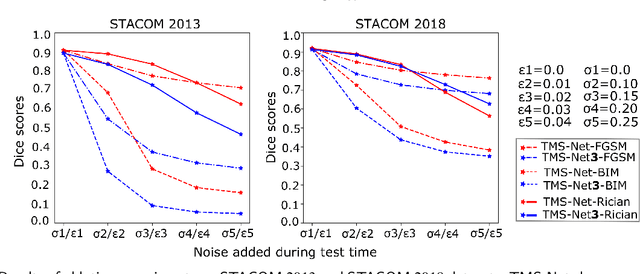
Abstract:Recently, deep networks have shown impressive performance for the segmentation of cardiac Magnetic Resonance Imaging (MRI) images. However, their achievement is proving slow to transition to widespread use in medical clinics because of robustness issues leading to low trust of clinicians to their results. Predicting run-time quality of segmentation masks can be useful to warn clinicians against poor results. Despite its importance, there are few studies on this problem. To address this gap, we propose a quality control method based on the agreement across decoders of a multi-view network, TMS-Net, measured by the cosine similarity. The network takes three view inputs resliced from the same 3D image along different axes. Different from previous multi-view networks, TMS-Net has a single encoder and three decoders, leading to better noise robustness, segmentation performance and run-time quality estimation in our experiments on the segmentation of the left atrium on STACOM 2013 and STACOM 2018 challenge datasets. We also present a way to generate poor segmentation masks by using noisy images generated with engineered noise and Rician noise to simulate undertraining, high anisotropy and poor imaging settings problems. Our run-time quality estimation method show a good classification of poor and good quality segmentation masks with an AUC reaching to 0.97 on STACOM 2018. We believe that TMS-Net and our run-time quality estimation method has a high potential to increase the thrust of clinicians to automatic image analysis tools.
Estimating Cardiac Tissue Conductivity from Electrograms with Fully Convolutional Networks
Dec 06, 2022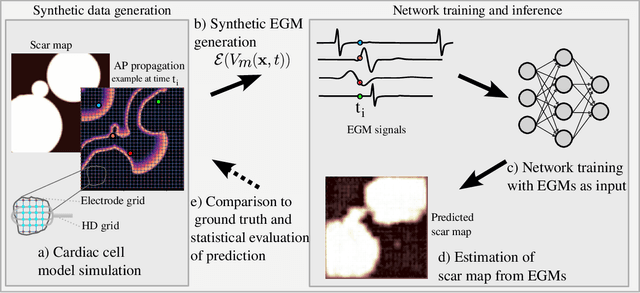
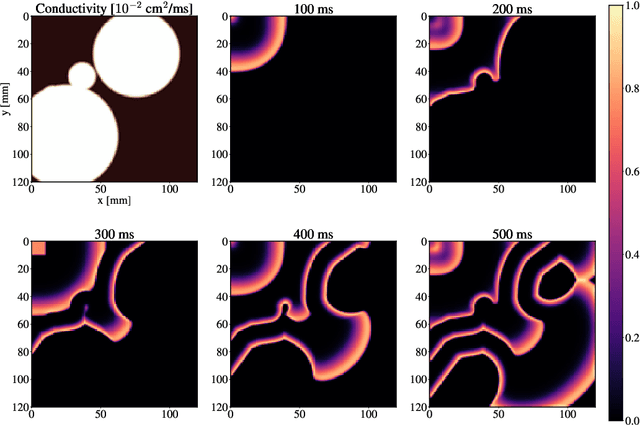
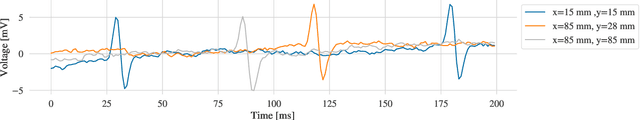
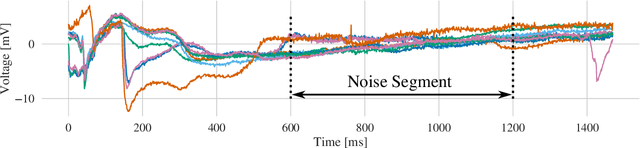
Abstract:Atrial Fibrillation (AF) is characterized by disorganised electrical activity in the atria and is known to be sustained by the presence of regions of fibrosis (scars) or functional cellular remodeling, both of which may lead to areas of slow conduction. Estimating the effective conductivity of the myocardium and identifying regions of abnormal propagation is therefore crucial for the effective treatment of AF. We hypothesise that the spatial distribution of tissue conductivity can be directly inferred from an array of concurrently acquired contact electrograms (EGMs). We generate a dataset of simulated cardiac AP propagation using randomised scar distributions and a phenomenological cardiac model and calculate contact electrograms at various positions on the field. A deep neural network, based on a modified U-net architecture, is trained to estimate the location of the scar and quantify conductivity of the tissue with a Jaccard index of $91$%. We adapt a wavelet-based surrogate testing analysis to confirm that the inferred conductivity distribution is an accurate representation of the ground truth input to the model. We find that the root mean square error (RMSE) between the ground truth and our predictions is significantly smaller ($p_{val}=0.007$) than the RMSE between the ground truth and surrogate samples.
REMuS-GNN: A Rotation-Equivariant Model for Simulating Continuum Dynamics
May 05, 2022
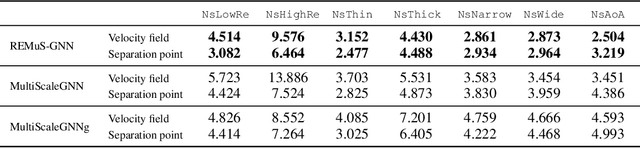
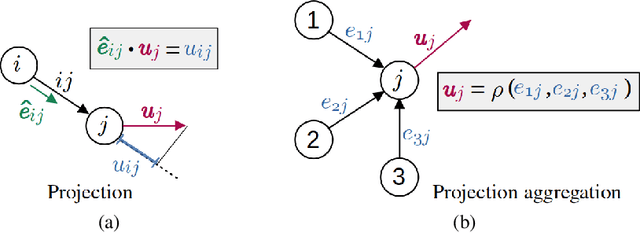
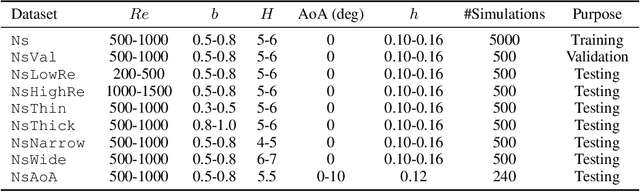
Abstract:Numerical simulation is an essential tool in many areas of science and engineering, but its performance often limits application in practice or when used to explore large parameter spaces. On the other hand, surrogate deep learning models, while accelerating simulations, often exhibit poor accuracy and ability to generalise. In order to improve these two factors, we introduce REMuS-GNN, a rotation-equivariant multi-scale model for simulating continuum dynamical systems encompassing a range of length scales. REMuS-GNN is designed to predict an output vector field from an input vector field on a physical domain discretised into an unstructured set of nodes. Equivariance to rotations of the domain is a desirable inductive bias that allows the network to learn the underlying physics more efficiently, leading to improved accuracy and generalisation compared with similar architectures that lack such symmetry. We demonstrate and evaluate this method on the incompressible flow around elliptical cylinders.
Towards Fast Simulation of Environmental Fluid Mechanics with Multi-Scale Graph Neural Networks
May 05, 2022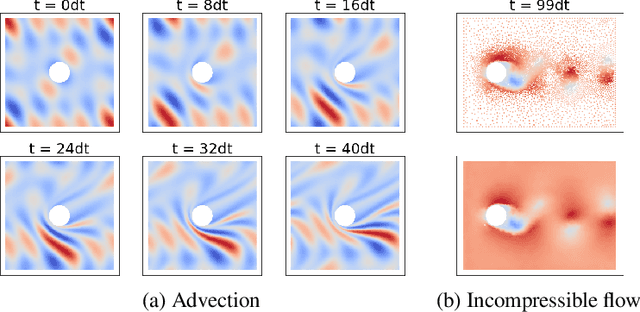

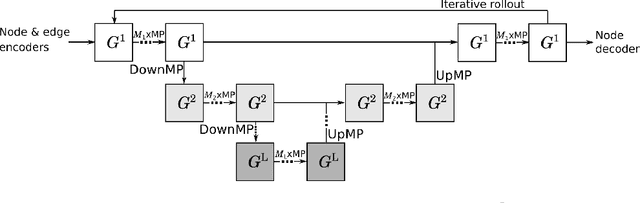
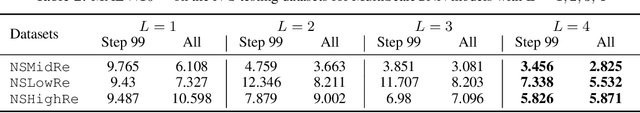
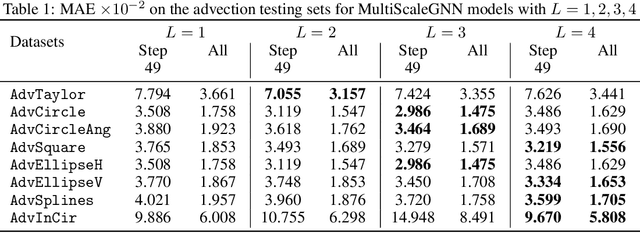
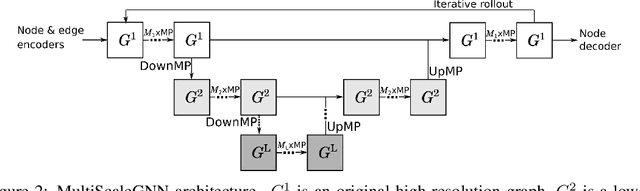
Abstract:Numerical simulators are essential tools in the study of natural fluid-systems, but their performance often limits application in practice. Recent machine-learning approaches have demonstrated their ability to accelerate spatio-temporal predictions, although, with only moderate accuracy in comparison. Here we introduce MultiScaleGNN, a novel multi-scale graph neural network model for learning to infer unsteady continuum mechanics in problems encompassing a range of length scales and complex boundary geometries. We demonstrate this method on advection problems and incompressible fluid dynamics, both fundamental phenomena in oceanic and atmospheric processes. Our results show good extrapolation to new domain geometries and parameters for long-term temporal simulations. Simulations obtained with MultiScaleGNN are between two and four orders of magnitude faster than those on which it was trained.
Tempera: Spatial Transformer Feature Pyramid Network for Cardiac MRI Segmentation
Mar 01, 2022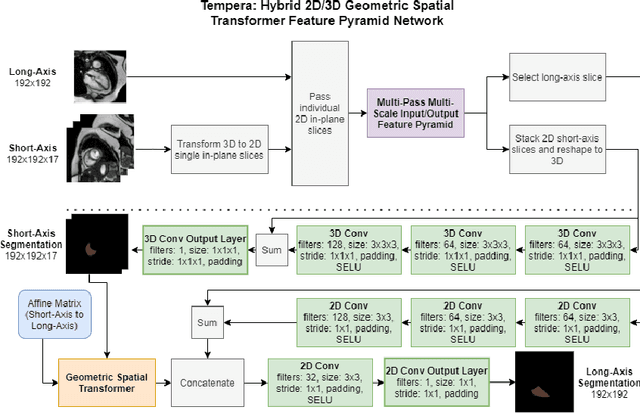
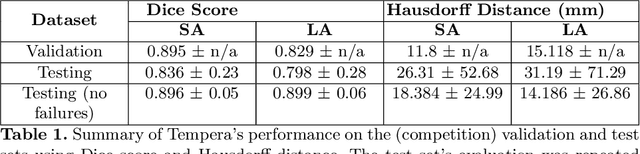
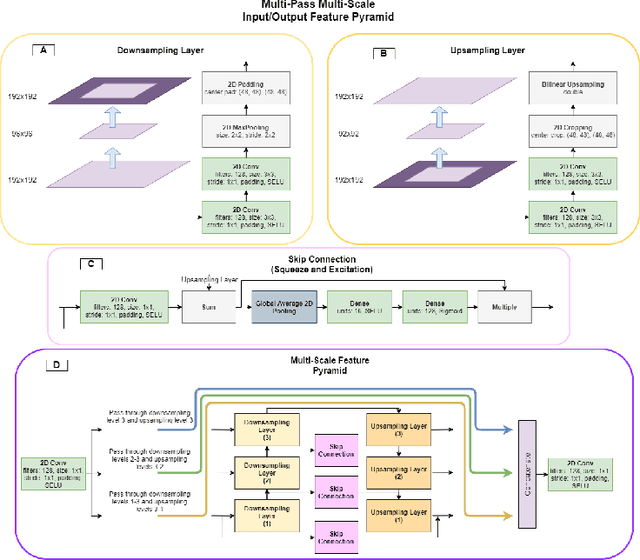
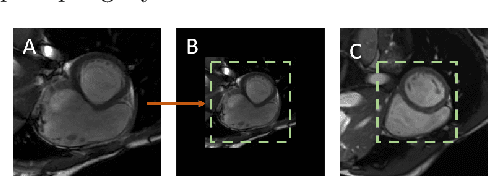
Abstract:Assessing the structure and function of the right ventricle (RV) is important in the diagnosis of several cardiac pathologies. However, it remains more challenging to segment the RV than the left ventricle (LV). In this paper, we focus on segmenting the RV in both short (SA) and long-axis (LA) cardiac MR images simultaneously. For this task, we propose a new multi-input/output architecture, hybrid 2D/3D geometric spatial TransformEr Multi-Pass fEature pyRAmid (Tempera). Our feature pyramid extends current designs by allowing not only a multi-scale feature output but multi-scale SA and LA input images as well. Tempera transfers learned features between SA and LA images via layer weight sharing and incorporates a geometric target transformer to map the predicted SA segmentation to LA space. Our model achieves an average Dice score of 0.836 and 0.798 for the SA and LA, respectively, and 26.31 mm and 31.19 mm Hausdorff distances. This opens up the potential for the incorporation of RV segmentation models into clinical workflows.
Simulating Continuum Mechanics with Multi-Scale Graph Neural Networks
Jun 09, 2021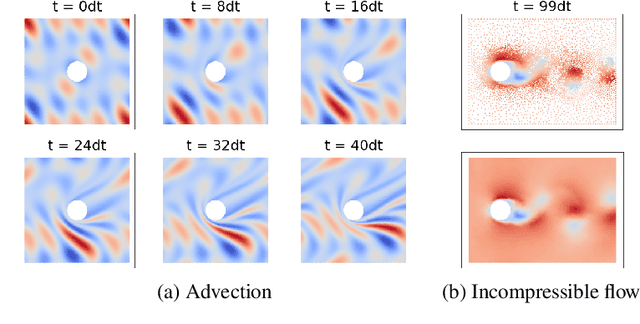

Abstract:Continuum mechanics simulators, numerically solving one or more partial differential equations, are essential tools in many areas of science and engineering, but their performance often limits application in practice. Recent modern machine learning approaches have demonstrated their ability to accelerate spatio-temporal predictions, although, with only moderate accuracy in comparison. Here we introduce MultiScaleGNN, a novel multi-scale graph neural network model for learning to infer unsteady continuum mechanics. MultiScaleGNN represents the physical domain as an unstructured set of nodes, and it constructs one or more graphs, each of them encoding different scales of spatial resolution. Successive learnt message passing between these graphs improves the ability of GNNs to capture and forecast the system state in problems encompassing a range of length scales. Using graph representations, MultiScaleGNN can impose periodic boundary conditions as an inductive bias on the edges in the graphs, and achieve independence to the nodes' positions. We demonstrate this method on advection problems and incompressible fluid dynamics. Our results show that the proposed model can generalise from uniform advection fields to high-gradient fields on complex domains at test time and infer long-term Navier-Stokes solutions within a range of Reynolds numbers. Simulations obtained with MultiScaleGNN are between two and four orders of magnitude faster than the ones on which it was trained.
Comparing recurrent and convolutional neural networks for predicting wave propagation
Mar 09, 2020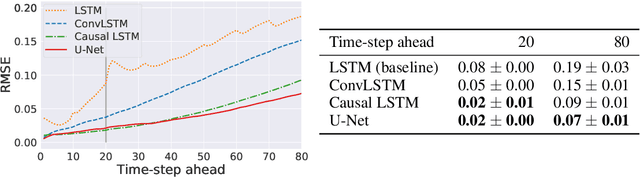
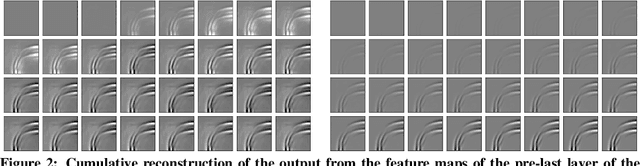
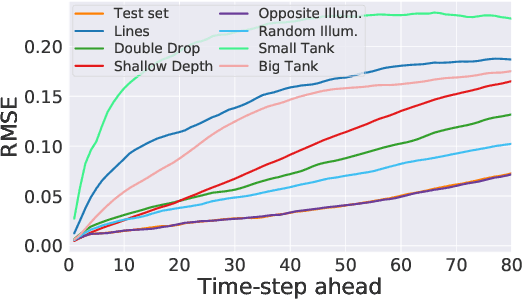
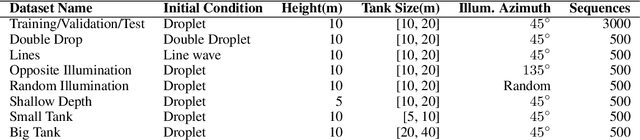
Abstract:Dynamical systems can be modelled by partial differential equations and numerical computations are used everywhere in science and engineering. In this work, we investigate the performance of recurrent and convolutional deep neural network architectures to predict the surface waves. The system is governed by the Saint-Venant equations. We improve on the long-term prediction over previous methods while keeping the inference time at a fraction of numerical simulations. We also show that convolutional networks perform at least as well as recurrent networks in this task. Finally, we assess the generalisation capability of each network by extrapolating in longer time-frames and in different physical settings.
Approximating the solution to wave propagation using deep neural networks
Dec 04, 2018
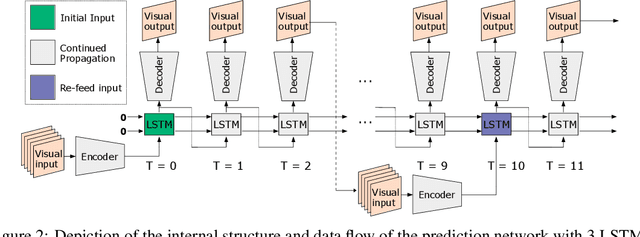
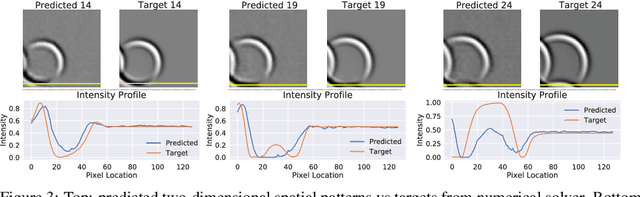
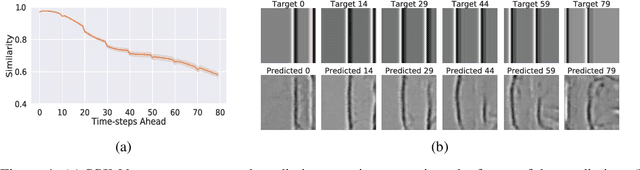
Abstract:Humans gain an implicit understanding of physical laws through observing and interacting with the world. Endowing an autonomous agent with an understanding of physical laws through experience and observation is seldom practical: we should seek alternatives. Fortunately, many of the laws of behaviour of the physical world can be derived from prior knowledge of dynamical systems, expressed through the use of partial differential equations. In this work, we suggest a neural network capable of understanding a specific physical phenomenon: wave propagation in a two-dimensional medium. We define `understanding' in this context as the ability to predict the future evolution of the spatial patterns of rendered wave amplitude from a relatively small set of initial observations. The inherent complexity of the wave equations -- together with the existence of reflections and interference -- makes the prediction problem non-trivial. A network capable of making approximate predictions also unlocks the opportunity to speed-up numerical simulations for wave propagation. To this aim, we created a novel dataset of simulated wave motion and built a predictive deep neural network comprising of three main blocks: an encoder, a propagator made by 3 LSTMs, and a decoder. Results show reasonable predictions for as long as 80 time steps into the future on a dataset not seen during training. Furthermore, the network is able to generalize to an initial condition that is qualitatively different from those seen during training.
Rethinking multiscale cardiac electrophysiology with machine learning and predictive modelling
Oct 09, 2018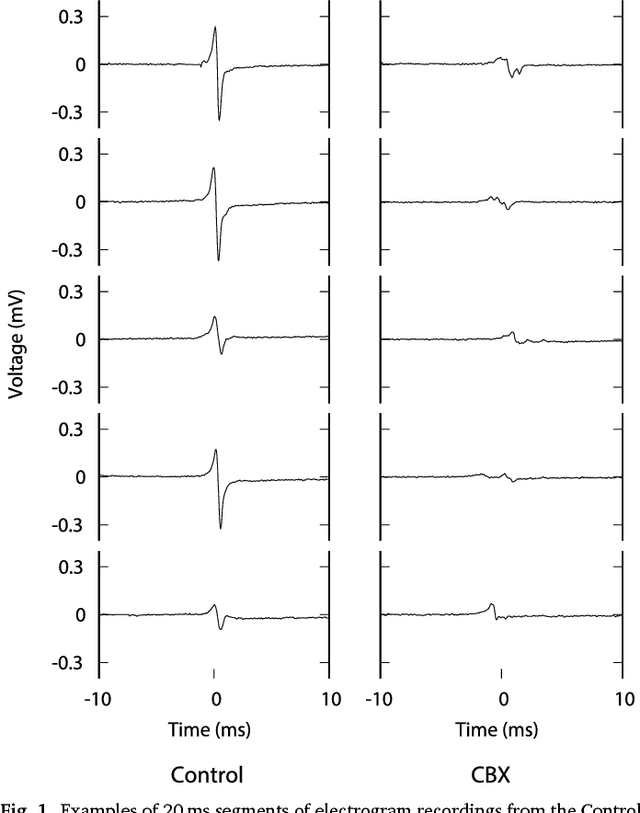
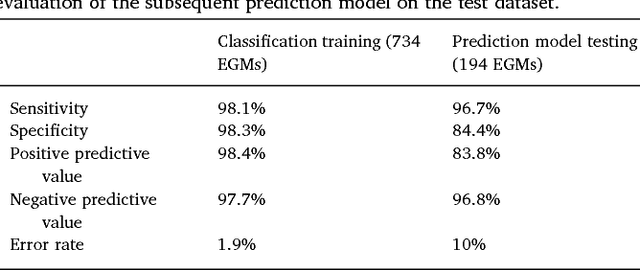
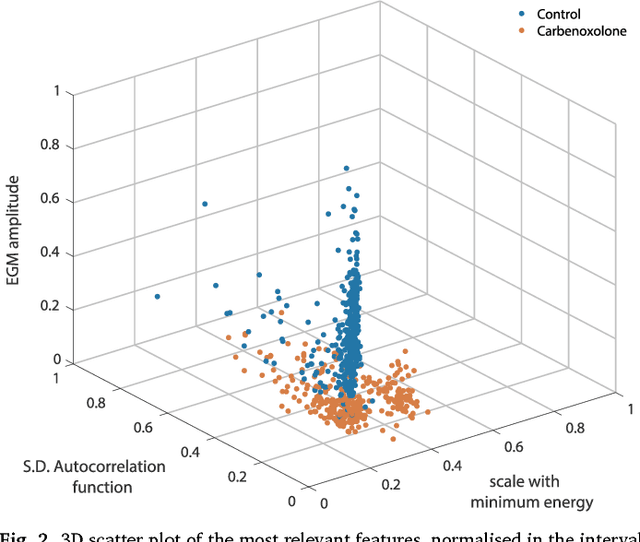
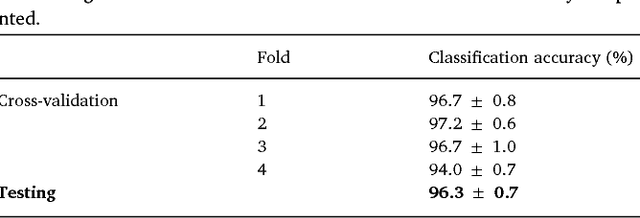
Abstract:We review some of the latest approaches to analysing cardiac electrophysiology data using machine learning and predictive modelling. Cardiac arrhythmias, particularly atrial fibrillation, are a major global healthcare challenge. Treatment is often through catheter ablation, which involves the targeted localized destruction of regions of the myocardium responsible for initiating or perpetuating the arrhythmia. Ablation targets are either anatomically defined, or identified based on their functional properties as determined through the analysis of contact intracardiac electrograms acquired with increasing spatial density by modern electroanatomic mapping systems. While numerous quantitative approaches have been investigated over the past decades for identifying these critical curative sites, few have provided a reliable and reproducible advance in success rates. Machine learning techniques, including recent deep-learning approaches, offer a potential route to gaining new insight from this wealth of highly complex spatio-temporal information that existing methods struggle to analyse. Coupled with predictive modelling, these techniques offer exciting opportunities to advance the field and produce more accurate diagnoses and robust personalised treatment. We outline some of these methods and illustrate their use in making predictions from the contact electrogram and augmenting predictive modelling tools, both by more rapidly predicting future states of the system and by inferring the parameters of these models from experimental observations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge