Fatmatulzehra Uslu
TMS-Net: A Segmentation Network Coupled With A Run-time Quality Control Method For Robust Cardiac Image Segmentation
Dec 21, 2022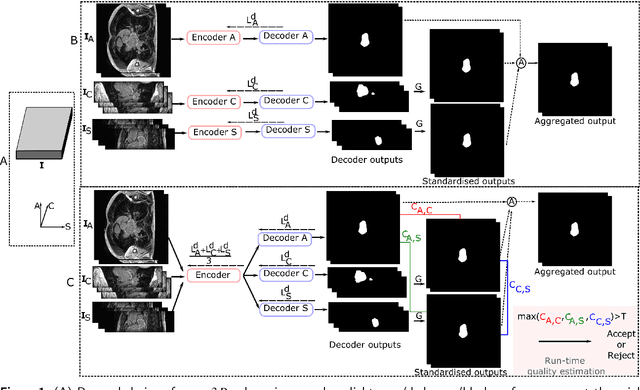

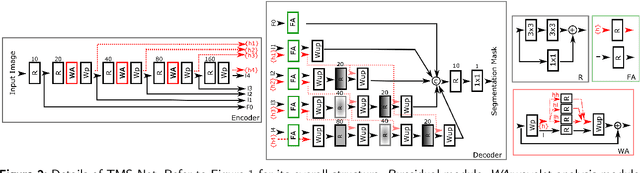
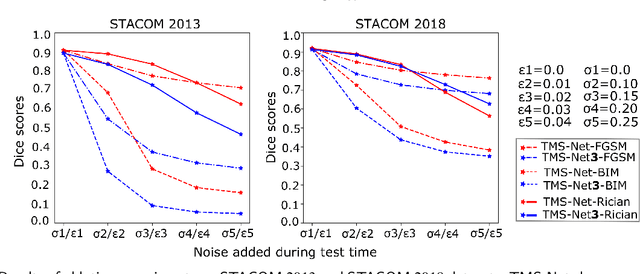
Abstract:Recently, deep networks have shown impressive performance for the segmentation of cardiac Magnetic Resonance Imaging (MRI) images. However, their achievement is proving slow to transition to widespread use in medical clinics because of robustness issues leading to low trust of clinicians to their results. Predicting run-time quality of segmentation masks can be useful to warn clinicians against poor results. Despite its importance, there are few studies on this problem. To address this gap, we propose a quality control method based on the agreement across decoders of a multi-view network, TMS-Net, measured by the cosine similarity. The network takes three view inputs resliced from the same 3D image along different axes. Different from previous multi-view networks, TMS-Net has a single encoder and three decoders, leading to better noise robustness, segmentation performance and run-time quality estimation in our experiments on the segmentation of the left atrium on STACOM 2013 and STACOM 2018 challenge datasets. We also present a way to generate poor segmentation masks by using noisy images generated with engineered noise and Rician noise to simulate undertraining, high anisotropy and poor imaging settings problems. Our run-time quality estimation method show a good classification of poor and good quality segmentation masks with an AUC reaching to 0.97 on STACOM 2018. We believe that TMS-Net and our run-time quality estimation method has a high potential to increase the thrust of clinicians to automatic image analysis tools.
An Inception Inspired Deep Network to Analyse Fundus Images
Nov 20, 2019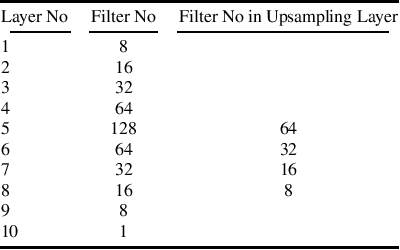
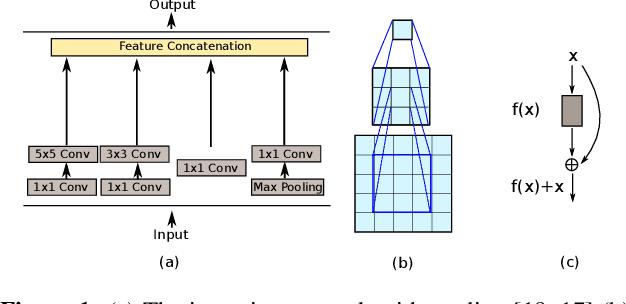
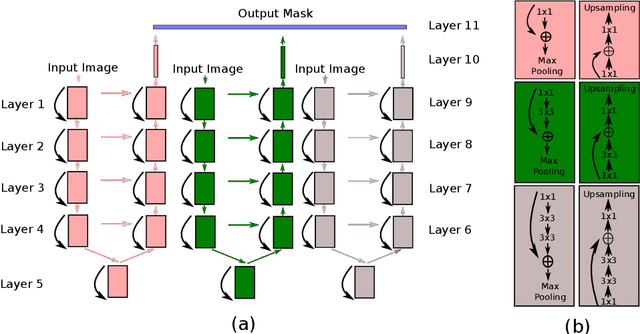
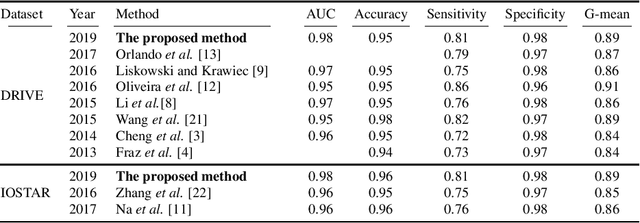
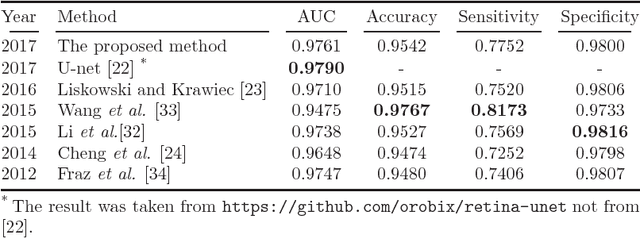
Abstract:A fundus image usually contains the optic disc, pathologies and other structures in addition to vessels to be segmented. This study proposes a deep network for vessel segmentation, whose architecture is inspired by inception modules. The network contains three sub-networks, each with a different filter size, which are connected in the last layer of the proposed network. According to experiments conducted in the DRIVE and IOSTAR, the performance of our network is found to be better than or comparable to that of the previous methods. We also observe that the sub-networks pay attention to different parts of an input image when producing an output map in the last layer of the proposed network; though, training of the proposed network is not constrained for this purpose.
A Recursive Bayesian Approach To Describe Retinal Vasculature Geometry
Nov 28, 2017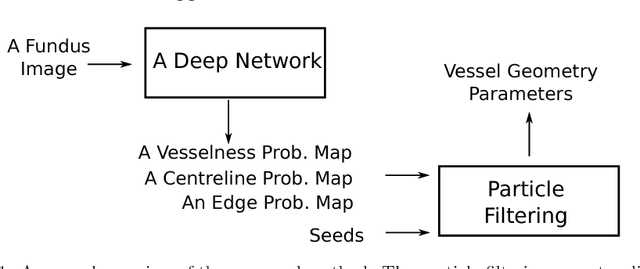

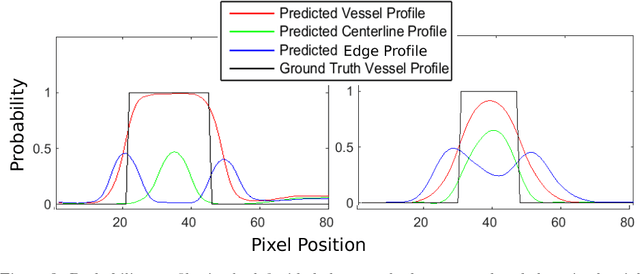
Abstract:Demographic studies suggest that changes in the retinal vasculature geometry, especially in vessel width, are associated with the incidence or progression of eye-related or systemic diseases. To date, the main information source for width estimation from fundus images has been the intensity profile between vessel edges. However, there are many factors affecting the intensity profile: pathologies, the central light reflex and local illumination levels, to name a few. In this study, we introduce three information sources for width estimation. These are the probability profiles of vessel interior, centreline and edge locations generated by a deep network. The probability profiles provide direct access to vessel geometry and are used in the likelihood calculation for a Bayesian method, particle filtering. We also introduce a geometric model which can handle non-ideal conditions of the probability profiles. Our experiments conducted on the REVIEW dataset yielded consistent estimates of vessel width, even in cases when one of the vessel edges is difficult to identify. Moreover, our results suggest that the method is better than human observers at locating edges of low contrast vessels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge