Fu Siong Ng
Explaining deep learning for ECG using time-localized clusters
Sep 18, 2025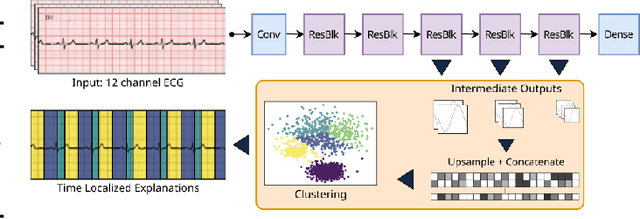
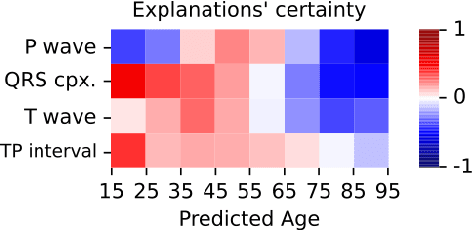
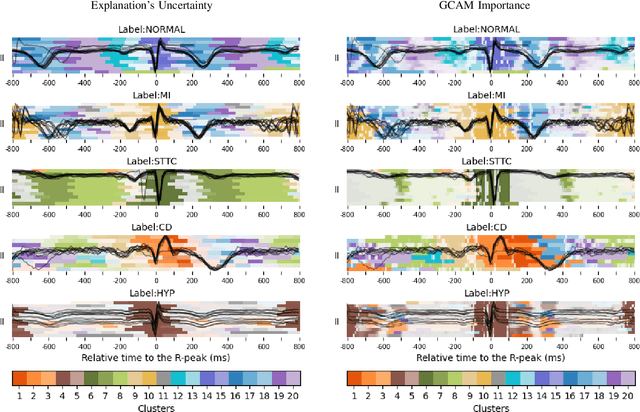
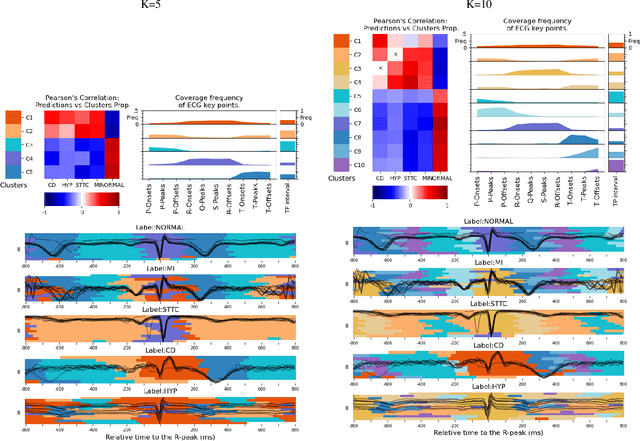
Abstract:Deep learning has significantly advanced electrocardiogram (ECG) analysis, enabling automatic annotation, disease screening, and prognosis beyond traditional clinical capabilities. However, understanding these models remains a challenge, limiting interpretation and gaining knowledge from these developments. In this work, we propose a novel interpretability method for convolutional neural networks applied to ECG analysis. Our approach extracts time-localized clusters from the model's internal representations, segmenting the ECG according to the learned characteristics while quantifying the uncertainty of these representations. This allows us to visualize how different waveform regions contribute to the model's predictions and assess the certainty of its decisions. By providing a structured and interpretable view of deep learning models for ECG, our method enhances trust in AI-driven diagnostics and facilitates the discovery of clinically relevant electrophysiological patterns.
Learning to Predict Global Atrial Fibrillation Dynamics from Sparse Measurements
Feb 13, 2025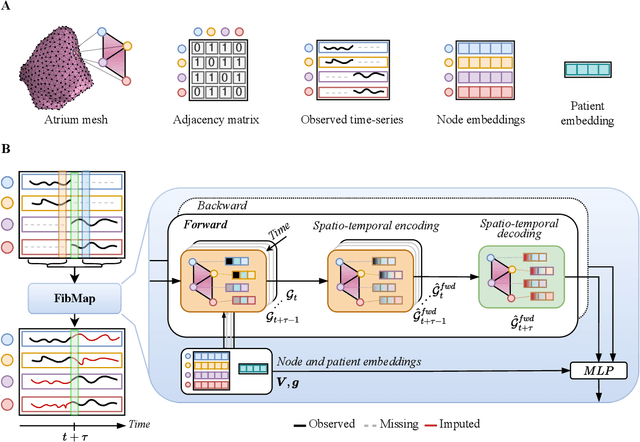
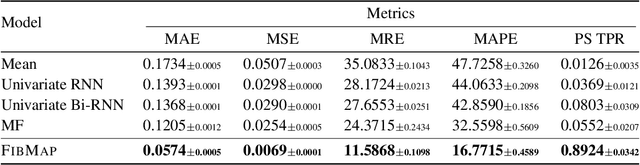
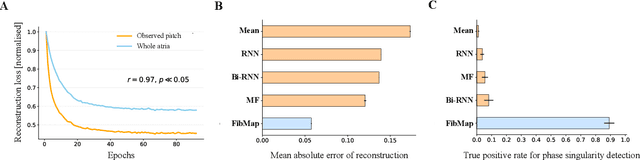
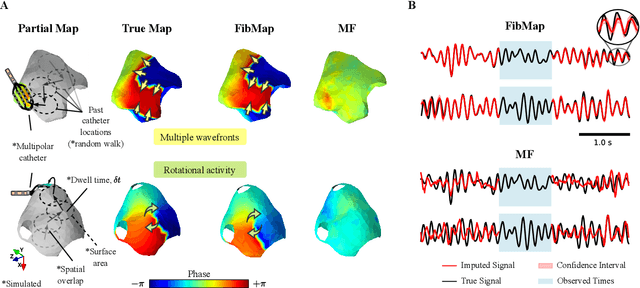
Abstract:Catheter ablation of Atrial Fibrillation (AF) consists of a one-size-fits-all treatment with limited success in persistent AF. This may be due to our inability to map the dynamics of AF with the limited resolution and coverage provided by sequential contact mapping catheters, preventing effective patient phenotyping for personalised, targeted ablation. Here we introduce FibMap, a graph recurrent neural network model that reconstructs global AF dynamics from sparse measurements. Trained and validated on 51 non-contact whole atria recordings, FibMap reconstructs whole atria dynamics from 10% surface coverage, achieving a 210% lower mean absolute error and an order of magnitude higher performance in tracking phase singularities compared to baseline methods. Clinical utility of FibMap is demonstrated on real-world contact mapping recordings, achieving reconstruction fidelity comparable to non-contact mapping. FibMap's state-spaces and patient-specific parameters offer insights for electrophenotyping AF. Integrating FibMap into clinical practice could enable personalised AF care and improve outcomes.
Online Graph Learning via Time-Vertex Adaptive Filters: From Theory to Cardiac Fibrillation
Nov 03, 2024Abstract:Graph Signal Processing (GSP) provides a powerful framework for analysing complex, interconnected systems by modelling data as signals on graphs. Recent advances in GSP have enabled the learning of graph structures from observed signals, but these methods often struggle with time-varying systems and real-time applications. Adaptive filtering techniques, while effective for online learning, have seen limited application in graph topology estimation from a GSP perspective. To this end, we introduce AdaCGP, an online algorithm for adaptive estimation of the Graph Shift Operator (GSO) from multivariate time series. The GSO is estimated from an adaptive time-vertex autoregressive model through recursive update formulae designed to address sparsity, shift-invariance and bias. Through simulations, we show that AdaCGP performs consistently well across various graph topologies, and achieves improvements in excess of 82% for GSO estimation compared to baseline adaptive vector autoregressive models. In addition, our online variable splitting approach for enforcing sparsity enables near-perfect precision in identifying causal connections while maintaining low false positive rates upon optimisation of the forecast error. Finally, AdaCGP's ability to track changes in graph structure is demonstrated on recordings of ventricular fibrillation dynamics in response to an anti-arrhythmic drug. AdaCGP is shown to be able to identify the stability of critical conduction patterns that may be maintaining the arrhythmia in an intuitive way, together with its potential to support diagnosis and treatment strategies.
Improving Diffusion Models for ECG Imputation with an Augmented Template Prior
Oct 24, 2023



Abstract:Pulsative signals such as the electrocardiogram (ECG) are extensively collected as part of routine clinical care. However, noisy and poor-quality recordings, leading to missing values, are a major issue for signals collected using mobile health systems, decreasing the signal quality and affecting the automated downstream tasks. Recent studies have explored imputation of missing values for ECG with probabilistic time-series models. Nevertheless, in comparison with the deterministic models, their performance is still limited, as the variations across subjects and heart-beat relationships are not explicitly considered in the training objective. In this work, to improve the ECG imputation and forecasting accuracy with probabilistic models, we present an template-guided denoising diffusion probabilistic model, PulseDiff, which is conditioned an informative prior for a range of health conditions. Specifically, 1) we first extract a subject-level pulsative template from the observation as an informative prior of missing values, which captures the personal characteristics; 2) we then add beat-level stochastic shift terms on the template for prior augmentation, which considers the beat-level variance of positioning and amplitude; 3) we finally design a confidence score to consider the health condition of subject, which ensures our prior is provided in a safe way. Experiments with the PTBXL dataset reveal PulseDiff improves the performance of two strong DDPMs baseline models, CSDI and SSSD$^{S4}$, verifying our method guides the generation of DDPMs while managing the uncertainty. When combining with SSSD$^{S4}$, our PulseDiff method outperforms the leading deterministic model for short-interval missing data and is comparable for long-interval data loss.
Rethinking multiscale cardiac electrophysiology with machine learning and predictive modelling
Oct 09, 2018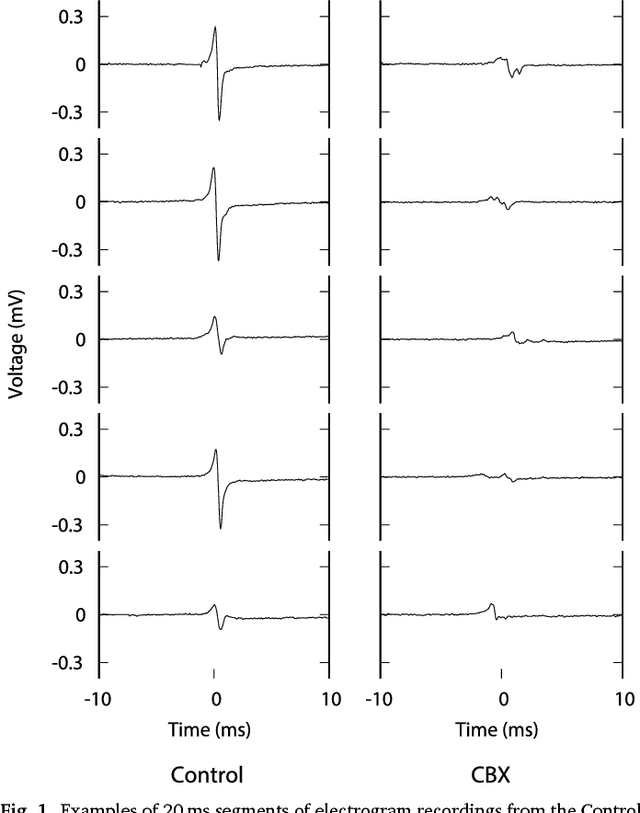
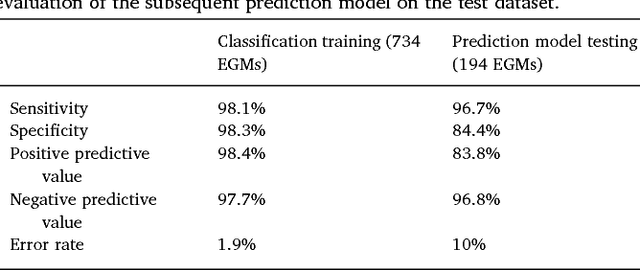
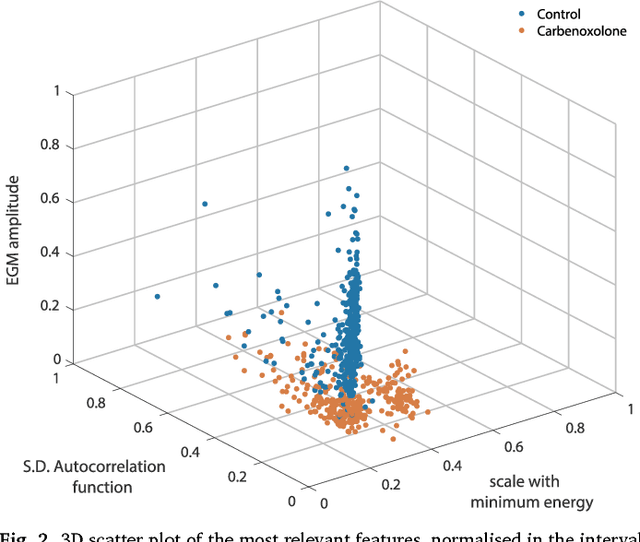
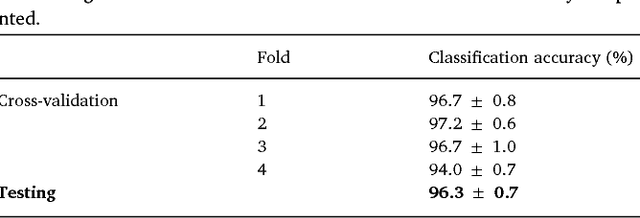
Abstract:We review some of the latest approaches to analysing cardiac electrophysiology data using machine learning and predictive modelling. Cardiac arrhythmias, particularly atrial fibrillation, are a major global healthcare challenge. Treatment is often through catheter ablation, which involves the targeted localized destruction of regions of the myocardium responsible for initiating or perpetuating the arrhythmia. Ablation targets are either anatomically defined, or identified based on their functional properties as determined through the analysis of contact intracardiac electrograms acquired with increasing spatial density by modern electroanatomic mapping systems. While numerous quantitative approaches have been investigated over the past decades for identifying these critical curative sites, few have provided a reliable and reproducible advance in success rates. Machine learning techniques, including recent deep-learning approaches, offer a potential route to gaining new insight from this wealth of highly complex spatio-temporal information that existing methods struggle to analyse. Coupled with predictive modelling, these techniques offer exciting opportunities to advance the field and produce more accurate diagnoses and robust personalised treatment. We outline some of these methods and illustrate their use in making predictions from the contact electrogram and augmenting predictive modelling tools, both by more rapidly predicting future states of the system and by inferring the parameters of these models from experimental observations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge