Stefan Kachel
Department of Radiology, The University of Melbourne, Parkville, Department of Radiology, Columbia University in the City of New York
A Review of Published Machine Learning Natural Language Processing Applications for Protocolling Radiology Imaging
Jun 23, 2022

Abstract:Machine learning (ML) is a subfield of Artificial intelligence (AI), and its applications in radiology are growing at an ever-accelerating rate. The most studied ML application is the automated interpretation of images. However, natural language processing (NLP), which can be combined with ML for text interpretation tasks, also has many potential applications in radiology. One such application is automation of radiology protocolling, which involves interpreting a clinical radiology referral and selecting the appropriate imaging technique. It is an essential task which ensures that the correct imaging is performed. However, the time that a radiologist must dedicate to protocolling could otherwise be spent reporting, communicating with referrers, or teaching. To date, there have been few publications in which ML models were developed that use clinical text to automate protocol selection. This article reviews the existing literature in this field. A systematic assessment of the published models is performed with reference to best practices suggested by machine learning convention. Progress towards implementing automated protocolling in a clinical setting is discussed.
CardiSort: a convolutional neural network for cross vendor automated sorting of cardiac MR images
Sep 17, 2021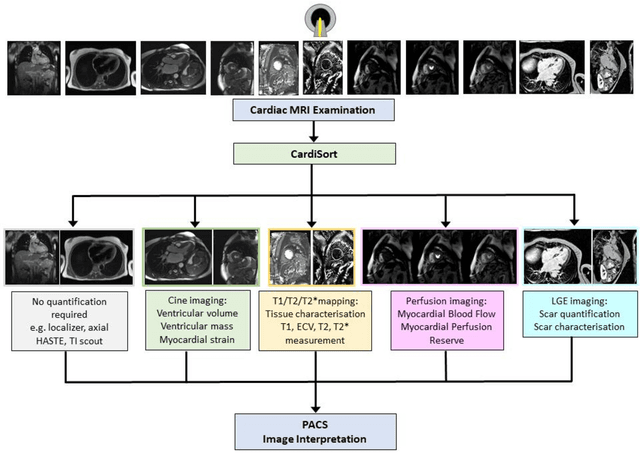
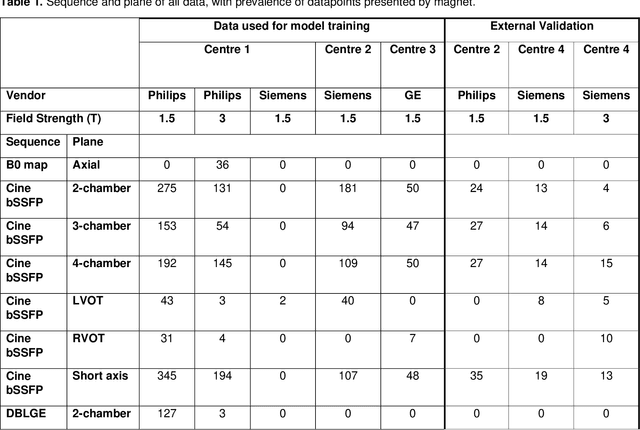
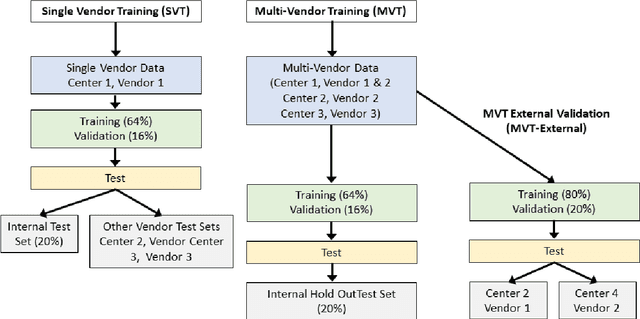
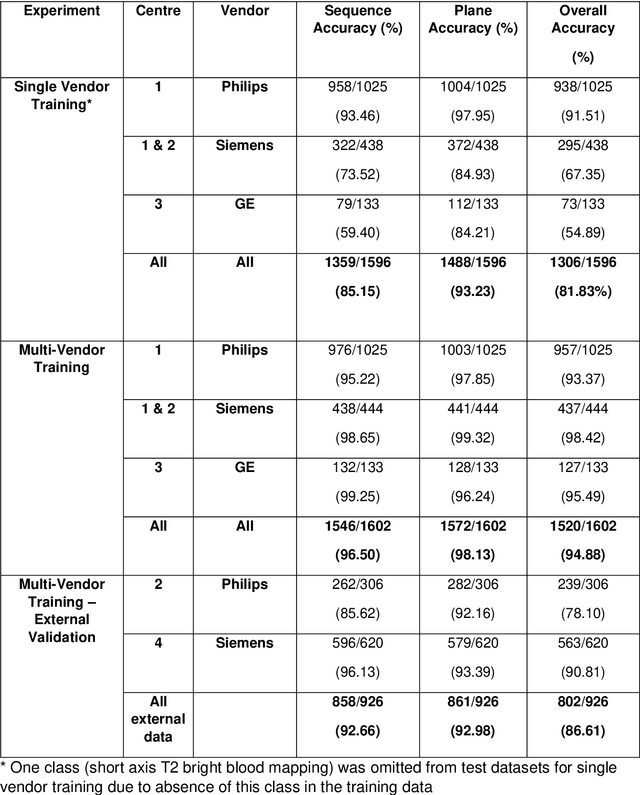
Abstract:Objectives: To develop an image-based automatic deep learning method to classify cardiac MR images by sequence type and imaging plane for improved clinical post-processing efficiency. Methods: Multi-vendor cardiac MRI studies were retrospectively collected from 4 centres and 3 vendors. A two-head convolutional neural network ('CardiSort') was trained to classify 35 sequences by imaging sequence (n=17) and plane (n=10). Single vendor training (SVT) on single centre images (n=234 patients) and multi-vendor training (MVT) with multicentre images (n = 479 patients, 3 centres) was performed. Model accuracy was compared to manual ground truth labels by an expert radiologist on a hold-out test set for both SVT and MVT. External validation of MVT (MVTexternal) was performed on data from 3 previously unseen magnet systems from 2 vendors (n=80 patients). Results: High sequence and plane accuracies were observed for SVT (85.2% and 93.2% respectively), and MVT (96.5% and 98.1% respectively) on the hold-out test set. MVTexternal yielded sequence accuracy of 92.7% and plane accuracy of 93.0%. There was high accuracy for common sequences and conventional cardiac planes. Poor accuracy was observed for underrepresented classes and sequences where there was greater variability in acquisition parameters across centres, such as perfusion imaging. Conclusions: A deep learning network was developed on multivendor data to classify MRI studies into component sequences and planes, with external validation. With refinement, it has potential to improve workflow by enabling automated sequence selection, an important first step in completely automated post-processing pipelines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge