Alistair A Young
Neural Implicit Heart Coordinates: 3D cardiac shape reconstruction from sparse segmentations
Dec 23, 2025Abstract:Accurate reconstruction of cardiac anatomy from sparse clinical images remains a major challenge in patient-specific modeling. While neural implicit functions have previously been applied to this task, their application to mapping anatomical consistency across subjects has been limited. In this work, we introduce Neural Implicit Heart Coordinates (NIHCs), a standardized implicit coordinate system, based on universal ventricular coordinates, that provides a common anatomical reference frame for the human heart. Our method predicts NIHCs directly from a limited number of 2D segmentations (sparse acquisition) and subsequently decodes them into dense 3D segmentations and high-resolution meshes at arbitrary output resolution. Trained on a large dataset of 5,000 cardiac meshes, the model achieves high reconstruction accuracy on clinical contours, with mean Euclidean surface errors of 2.51$\pm$0.33 mm in a diseased cohort (n=4549) and 2.3$\pm$0.36 mm in a healthy cohort (n=5576). The NIHC representation enables anatomically coherent reconstruction even under severe slice sparsity and segmentation noise, faithfully recovering complex structures such as the valve planes. Compared with traditional pipelines, inference time is reduced from over 60 s to 5-15 s. These results demonstrate that NIHCs constitute a robust and efficient anatomical representation for patient-specific 3D cardiac reconstruction from minimal input data.
Improved 3D Whole Heart Geometry from Sparse CMR Slices
Aug 14, 2024Abstract:Cardiac magnetic resonance (CMR) imaging and computed tomography (CT) are two common non-invasive imaging methods for assessing patients with cardiovascular disease. CMR typically acquires multiple sparse 2D slices, with unavoidable respiratory motion artefacts between slices, whereas CT acquires isotropic dense data but uses ionising radiation. In this study, we explore the combination of Slice Shifting Algorithm (SSA), Spatial Transformer Network (STN), and Label Transformer Network (LTN) to: 1) correct respiratory motion between segmented slices, and 2) transform sparse segmentation data into dense segmentation. All combinations were validated using synthetic motion-corrupted CMR slice segmentation generated from CT in 1699 cases, where the dense CT serves as the ground truth. In 199 testing cases, SSA-LTN achieved the best results for Dice score and Huasdorff distance (94.0% and 4.7 mm respectively, average over 5 labels) but gave topological errors in 8 cases. STN was effective as a plug-in tool for correcting all topological errors with minimal impact on overall performance (93.5% and 5.0 mm respectively). SSA also proves to be a valuable plug-in tool, enhancing performance over both STN-based and LTN-based models. The code for these different combinations is available at https://github.com/XESchong/STACOM2024.
Deep Learning Analysis of Cardiac MRI in Legacy Datasets: Multi-Ethnic Study of Atherosclerosis
Oct 28, 2021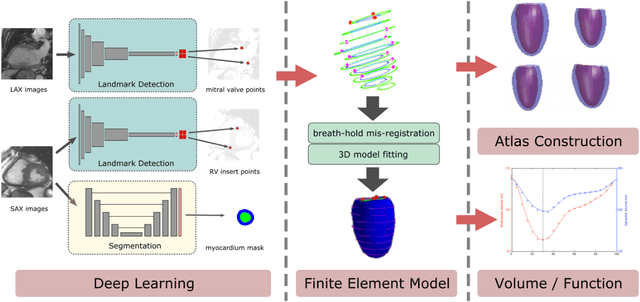
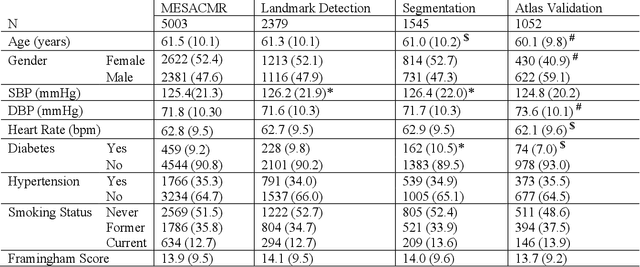
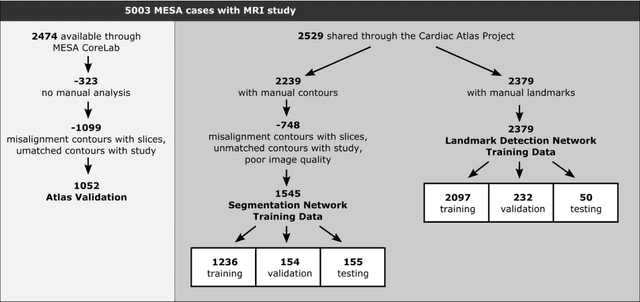
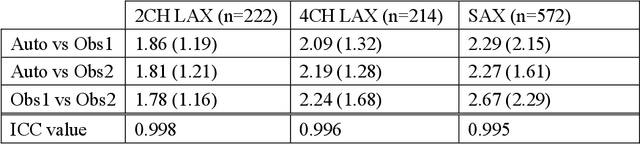
Abstract:The shape and motion of the heart provide essential clues to understanding the mechanisms of cardiovascular disease. With the advent of large-scale cardiac imaging data, statistical atlases become a powerful tool to provide automated and precise quantification of the status of patient-specific heart geometry with respect to reference populations. The Multi-Ethnic Study of Atherosclerosis (MESA), begun in 2000, was the first large cohort study to incorporate cardiovascular MRI in over 5000 participants, and there is now a wealth of follow-up data over 20 years. Building a machine learning based automated analysis is necessary to extract the additional imaging information necessary for expanding original manual analyses. However, machine learning tools trained on MRI datasets with different pulse sequences fail on such legacy datasets. Here, we describe an automated atlas construction pipeline using deep learning methods applied to the legacy cardiac MRI data in MESA. For detection of anatomical cardiac landmark points, a modified VGGNet convolutional neural network architecture was used in conjunction with a transfer learning sequence between two-chamber, four-chamber, and short-axis MRI views. A U-Net architecture was used for detection of the endocardial and epicardial boundaries in short axis images. Both network architectures resulted in good segmentation and landmark detection accuracies compared with inter-observer variations. Statistical relationships with common risk factors were similar between atlases derived from automated vs manual annotations. The automated atlas can be employed in future studies to examine the relationships between cardiac morphology and future events.
CardiSort: a convolutional neural network for cross vendor automated sorting of cardiac MR images
Sep 17, 2021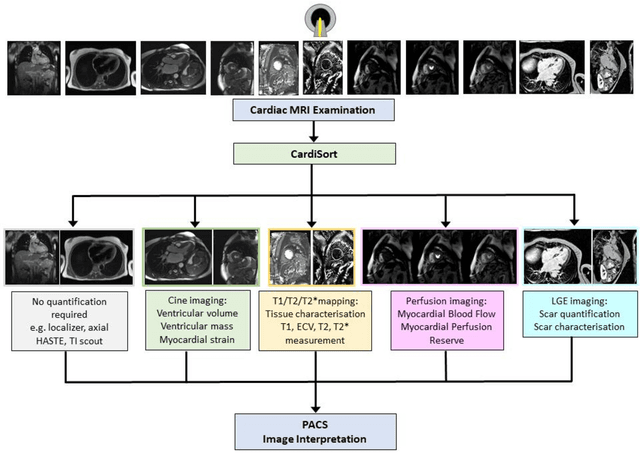
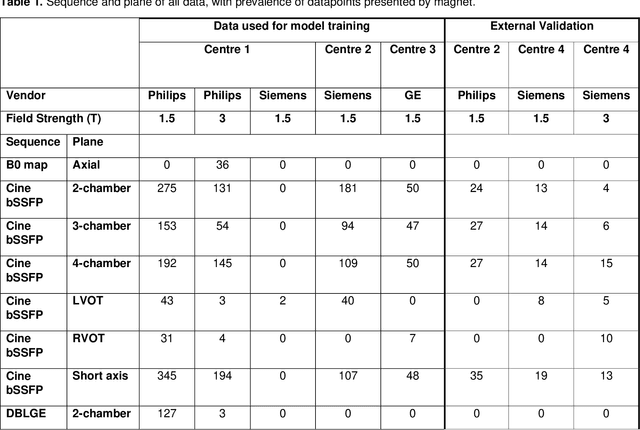
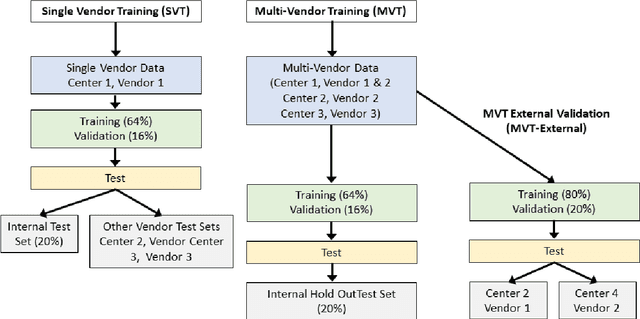
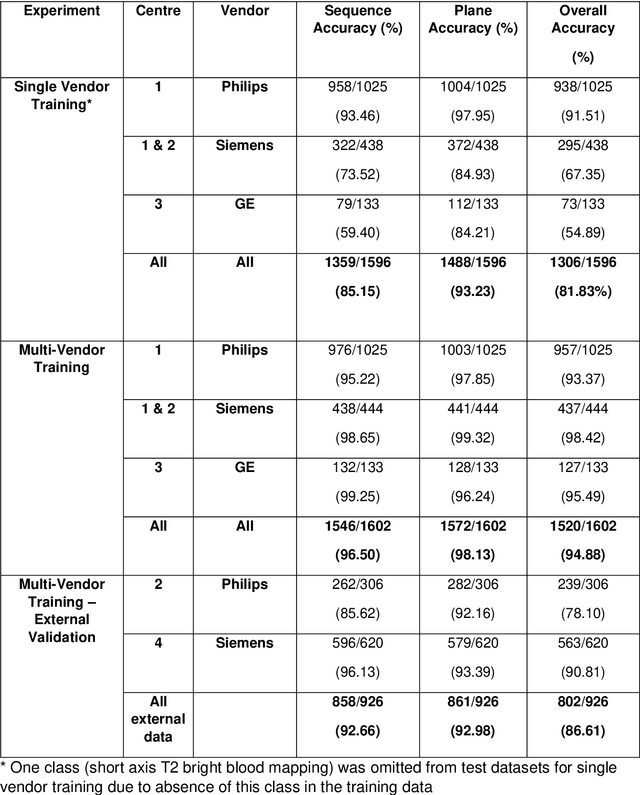
Abstract:Objectives: To develop an image-based automatic deep learning method to classify cardiac MR images by sequence type and imaging plane for improved clinical post-processing efficiency. Methods: Multi-vendor cardiac MRI studies were retrospectively collected from 4 centres and 3 vendors. A two-head convolutional neural network ('CardiSort') was trained to classify 35 sequences by imaging sequence (n=17) and plane (n=10). Single vendor training (SVT) on single centre images (n=234 patients) and multi-vendor training (MVT) with multicentre images (n = 479 patients, 3 centres) was performed. Model accuracy was compared to manual ground truth labels by an expert radiologist on a hold-out test set for both SVT and MVT. External validation of MVT (MVTexternal) was performed on data from 3 previously unseen magnet systems from 2 vendors (n=80 patients). Results: High sequence and plane accuracies were observed for SVT (85.2% and 93.2% respectively), and MVT (96.5% and 98.1% respectively) on the hold-out test set. MVTexternal yielded sequence accuracy of 92.7% and plane accuracy of 93.0%. There was high accuracy for common sequences and conventional cardiac planes. Poor accuracy was observed for underrepresented classes and sequences where there was greater variability in acquisition parameters across centres, such as perfusion imaging. Conclusions: A deep learning network was developed on multivendor data to classify MRI studies into component sequences and planes, with external validation. With refinement, it has potential to improve workflow by enabling automated sequence selection, an important first step in completely automated post-processing pipelines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge