David F. A. Lloyd
DCRA-Net: Attention-Enabled Reconstruction Model for Dynamic Fetal Cardiac MRI
Dec 19, 2024



Abstract:Dynamic fetal heart magnetic resonance imaging (MRI) presents unique challenges due to the fast heart rate of the fetus compared to adult subjects and uncontrolled fetal motion. This requires high temporal and spatial resolutions over a large field of view, in order to encompass surrounding maternal anatomy. In this work, we introduce Dynamic Cardiac Reconstruction Attention Network (DCRA-Net) - a novel deep learning model that employs attention mechanisms in spatial and temporal domains and temporal frequency representation of data to reconstruct the dynamics of the fetal heart from highly accelerated free-running (non-gated) MRI acquisitions. DCRA-Net was trained on retrospectively undersampled complex-valued cardiac MRIs from 42 fetal subjects and separately from 153 adult subjects, and evaluated on data from 14 fetal and 39 adult subjects respectively. Its performance was compared to L+S and k-GIN methods in both fetal and adult cases for an undersampling factor of 8x. The proposed network performed better than the comparators for both fetal and adult data, for both regular lattice and centrally weighted random undersampling. Aliased signals due to the undersampling were comprehensively resolved, and both the spatial details of the heart and its temporal dynamics were recovered with high fidelity. The highest performance was achieved when using lattice undersampling, data consistency and temporal frequency representation, yielding PSNR of 38 for fetal and 35 for adult cases. Our method is publicly available at https://github.com/denproc/DCRA-Net.
Multi-task learning for joint weakly-supervised segmentation and aortic arch anomaly classification in fetal cardiac MRI
Nov 13, 2023Abstract:Congenital Heart Disease (CHD) is a group of cardiac malformations present already during fetal life, representing the prevailing category of birth defects globally. Our aim in this study is to aid 3D fetal vessel topology visualisation in aortic arch anomalies, a group which encompasses a range of conditions with significant anatomical heterogeneity. We present a multi-task framework for automated multi-class fetal vessel segmentation from 3D black blood T2w MRI and anomaly classification. Our training data consists of binary manual segmentation masks of the cardiac vessels' region in individual subjects and fully-labelled anomaly-specific population atlases. Our framework combines deep learning label propagation using VoxelMorph with 3D Attention U-Net segmentation and DenseNet121 anomaly classification. We target 11 cardiac vessels and three distinct aortic arch anomalies, including double aortic arch, right aortic arch, and suspected coarctation of the aorta. We incorporate an anomaly classifier into our segmentation pipeline, delivering a multi-task framework with the primary motivation of correcting topological inaccuracies of the segmentation. The hypothesis is that the multi-task approach will encourage the segmenter network to learn anomaly-specific features. As a secondary motivation, an automated diagnosis tool may have the potential to enhance diagnostic confidence in a decision support setting. Our results showcase that our proposed training strategy significantly outperforms label propagation and a network trained exclusively on propagated labels. Our classifier outperforms a classifier trained exclusively on T2w volume images, with an average balanced accuracy of 0.99 (0.01) after joint training. Adding a classifier improves the anatomical and topological accuracy of all correctly classified double aortic arch subjects.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2023:015
Mutual Information-based Disentangled Neural Networks for Classifying Unseen Categories in Different Domains: Application to Fetal Ultrasound Imaging
Oct 30, 2020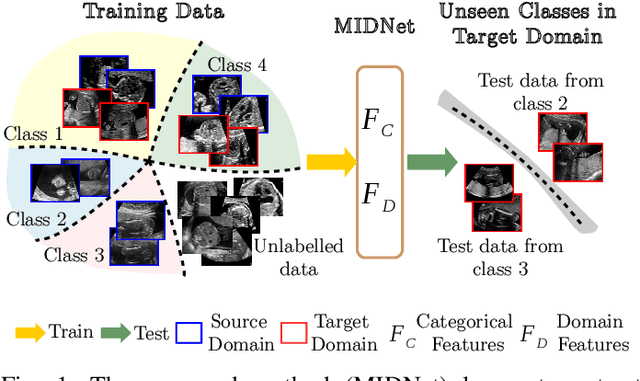
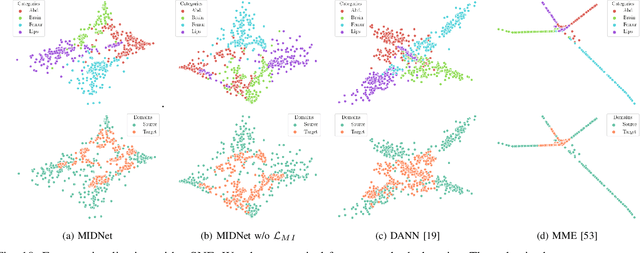
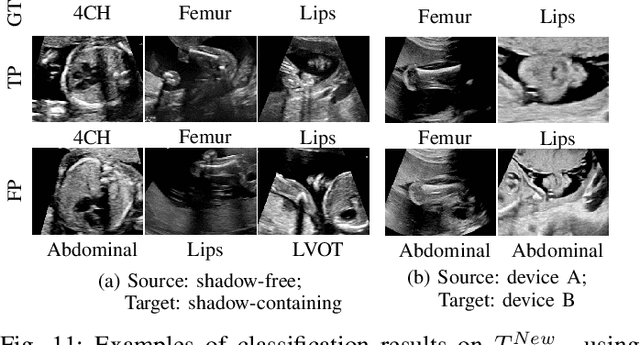
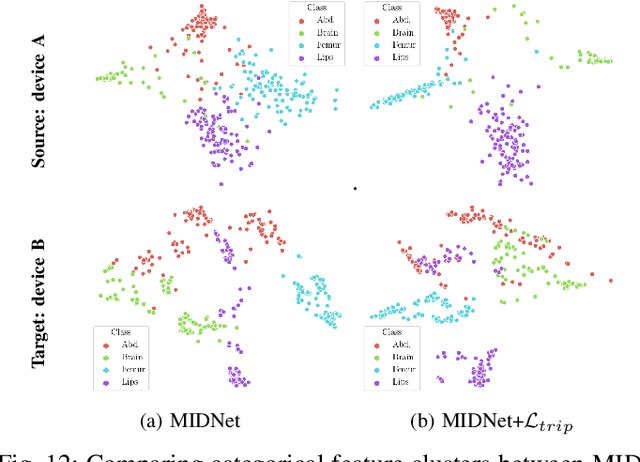
Abstract:Deep neural networks exhibit limited generalizability across images with different entangled domain features and categorical features. Learning generalizable features that can form universal categorical decision boundaries across domains is an interesting and difficult challenge. This problem occurs frequently in medical imaging applications when attempts are made to deploy and improve deep learning models across different image acquisition devices, across acquisition parameters or if some classes are unavailable in new training databases. To address this problem, we propose Mutual Information-based Disentangled Neural Networks (MIDNet), which extract generalizable categorical features to transfer knowledge to unseen categories in a target domain. The proposed MIDNet adopts a semi-supervised learning paradigm to alleviate the dependency on labeled data. This is important for real-world applications where data annotation is time-consuming, costly and requires training and expertise. We extensively evaluate the proposed method on fetal ultrasound datasets for two different image classification tasks where domain features are respectively defined by shadow artifacts and image acquisition devices. Experimental results show that the proposed method outperforms the state-of-the-art on the classification of unseen categories in a target domain with sparsely labeled training data.
PVR: Patch-to-Volume Reconstruction for Large Area Motion Correction of Fetal MRI
Nov 25, 2016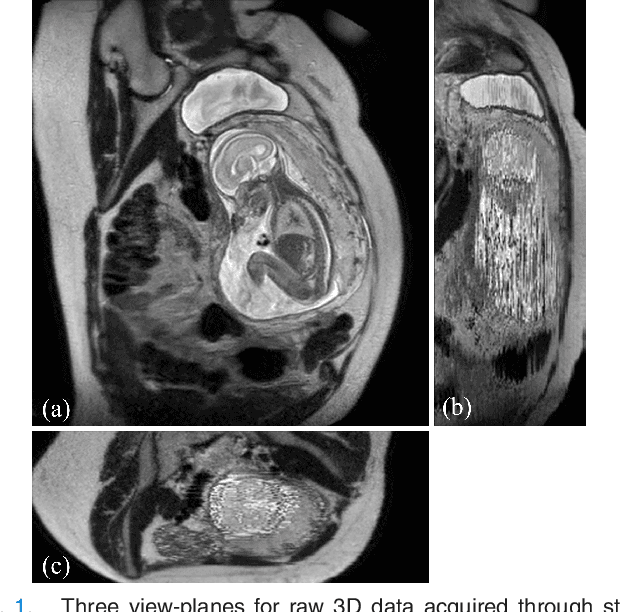
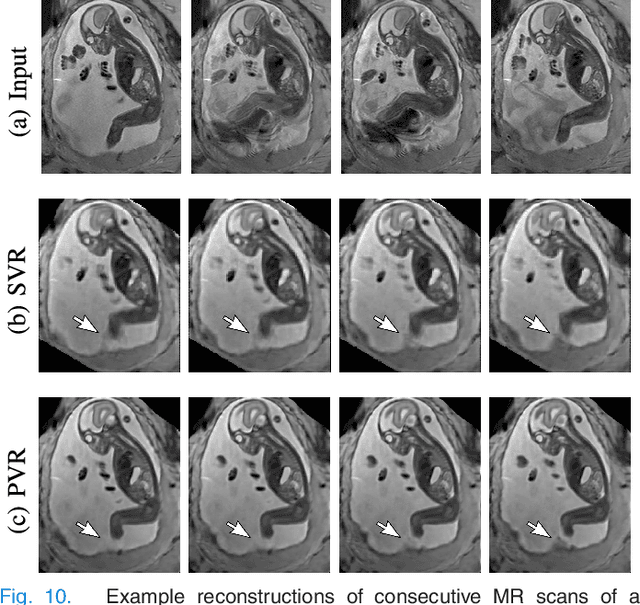
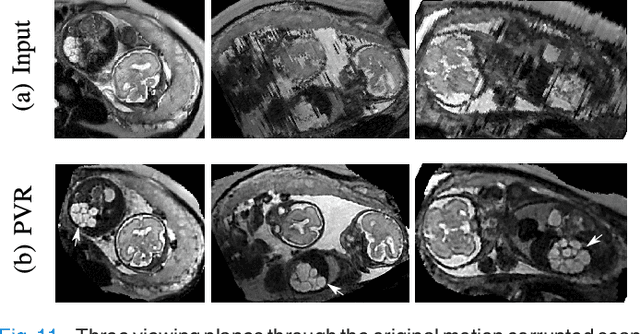
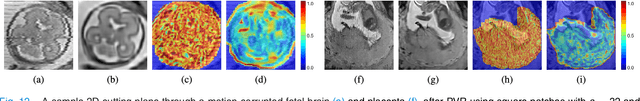
Abstract:In this paper we present a novel method for the correction of motion artifacts that are present in fetal Magnetic Resonance Imaging (MRI) scans of the whole uterus. Contrary to current slice-to-volume registration (SVR) methods, requiring an inflexible anatomical enclosure of a single investigated organ, the proposed patch-to-volume reconstruction (PVR) approach is able to reconstruct a large field of view of non-rigidly deforming structures. It relaxes rigid motion assumptions by introducing a specific amount of redundant information that is exploited with parallelized patch-wise optimization, super-resolution, and automatic outlier rejection. We further describe and provide an efficient parallel implementation of PVR allowing its execution within reasonable time on commercially available graphics processing units (GPU), enabling its use in the clinical practice. We evaluate PVR's computational overhead compared to standard methods and observe improved reconstruction accuracy in presence of affine motion artifacts of approximately 30% compared to conventional SVR in synthetic experiments. Furthermore, we have evaluated our method qualitatively and quantitatively on real fetal MRI data subject to maternal breathing and sudden fetal movements. We evaluate peak-signal-to-noise ratio (PSNR), structural similarity index (SSIM), and cross correlation (CC) with respect to the originally acquired data and provide a method for visual inspection of reconstruction uncertainty. With these experiments we demonstrate successful application of PVR motion compensation to the whole uterus, the human fetus, and the human placenta.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge