Alice Davidson
3D Reconstruction in Canonical Co-ordinate Space from Arbitrarily Oriented 2D Images
Jan 23, 2018



Abstract:Limited capture range, and the requirement to provide high quality initialization for optimization-based 2D/3D image registration methods, can significantly degrade the performance of 3D image reconstruction and motion compensation pipelines. Challenging clinical imaging scenarios, which contain significant subject motion such as fetal in-utero imaging, complicate the 3D image and volume reconstruction process. In this paper we present a learning based image registration method capable of predicting 3D rigid transformations of arbitrarily oriented 2D image slices, with respect to a learned canonical atlas co-ordinate system. Only image slice intensity information is used to perform registration and canonical alignment, no spatial transform initialization is required. To find image transformations we utilize a Convolutional Neural Network (CNN) architecture to learn the regression function capable of mapping 2D image slices to a 3D canonical atlas space. We extensively evaluate the effectiveness of our approach quantitatively on simulated Magnetic Resonance Imaging (MRI), fetal brain imagery with synthetic motion and further demonstrate qualitative results on real fetal MRI data where our method is integrated into a full reconstruction and motion compensation pipeline. Our learning based registration achieves an average spatial prediction error of 7 mm on simulated data and produces qualitatively improved reconstructions for heavily moving fetuses with gestational ages of approximately 20 weeks. Our model provides a general and computationally efficient solution to the 2D/3D registration initialization problem and is suitable for real-time scenarios.
Predicting Slice-to-Volume Transformation in Presence of Arbitrary Subject Motion
Mar 04, 2017



Abstract:This paper aims to solve a fundamental problem in intensity-based 2D/3D registration, which concerns the limited capture range and need for very good initialization of state-of-the-art image registration methods. We propose a regression approach that learns to predict rotation and translations of arbitrary 2D image slices from 3D volumes, with respect to a learned canonical atlas co-ordinate system. To this end, we utilize Convolutional Neural Networks (CNNs) to learn the highly complex regression function that maps 2D image slices into their correct position and orientation in 3D space. Our approach is attractive in challenging imaging scenarios, where significant subject motion complicates reconstruction performance of 3D volumes from 2D slice data. We extensively evaluate the effectiveness of our approach quantitatively on simulated MRI brain data with extreme random motion. We further demonstrate qualitative results on fetal MRI where our method is integrated into a full reconstruction and motion compensation pipeline. With our CNN regression approach we obtain an average prediction error of 7mm on simulated data, and convincing reconstruction quality of images of very young fetuses where previous methods fail. We further discuss applications to Computed Tomography and X-ray projections. Our approach is a general solution to the 2D/3D initialization problem. It is computationally efficient, with prediction times per slice of a few milliseconds, making it suitable for real-time scenarios.
PVR: Patch-to-Volume Reconstruction for Large Area Motion Correction of Fetal MRI
Nov 25, 2016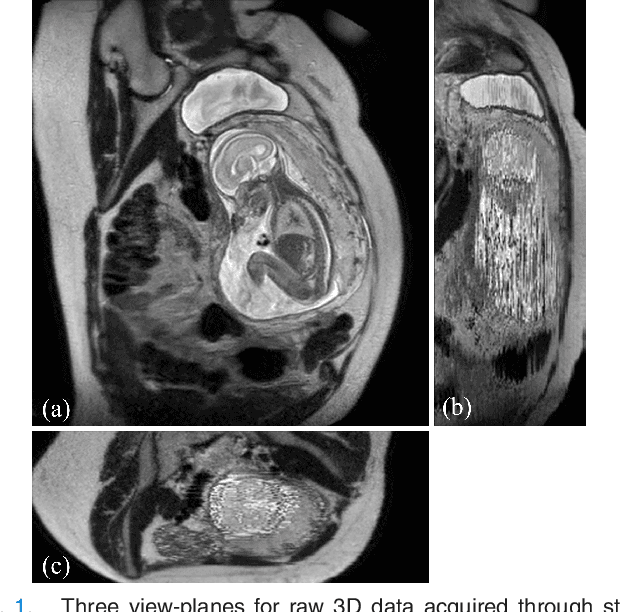
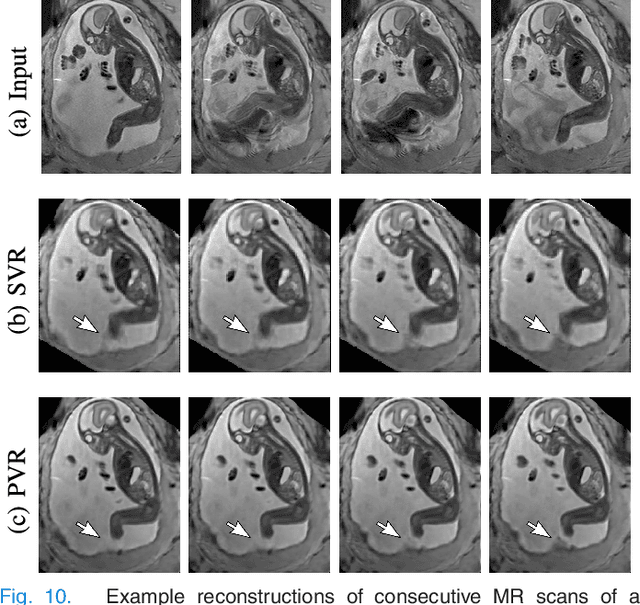
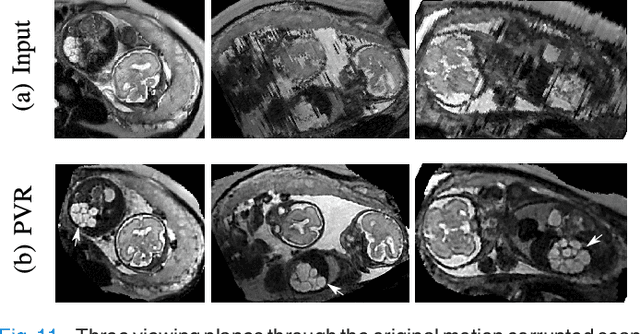
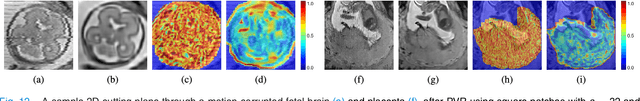
Abstract:In this paper we present a novel method for the correction of motion artifacts that are present in fetal Magnetic Resonance Imaging (MRI) scans of the whole uterus. Contrary to current slice-to-volume registration (SVR) methods, requiring an inflexible anatomical enclosure of a single investigated organ, the proposed patch-to-volume reconstruction (PVR) approach is able to reconstruct a large field of view of non-rigidly deforming structures. It relaxes rigid motion assumptions by introducing a specific amount of redundant information that is exploited with parallelized patch-wise optimization, super-resolution, and automatic outlier rejection. We further describe and provide an efficient parallel implementation of PVR allowing its execution within reasonable time on commercially available graphics processing units (GPU), enabling its use in the clinical practice. We evaluate PVR's computational overhead compared to standard methods and observe improved reconstruction accuracy in presence of affine motion artifacts of approximately 30% compared to conventional SVR in synthetic experiments. Furthermore, we have evaluated our method qualitatively and quantitatively on real fetal MRI data subject to maternal breathing and sudden fetal movements. We evaluate peak-signal-to-noise ratio (PSNR), structural similarity index (SSIM), and cross correlation (CC) with respect to the originally acquired data and provide a method for visual inspection of reconstruction uncertainty. With these experiments we demonstrate successful application of PVR motion compensation to the whole uterus, the human fetus, and the human placenta.
Learning under Distributed Weak Supervision
Jun 03, 2016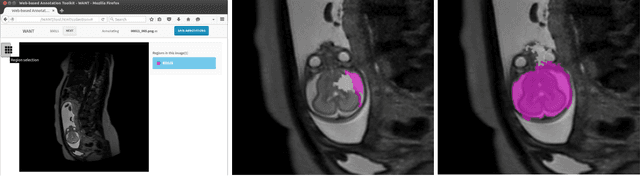
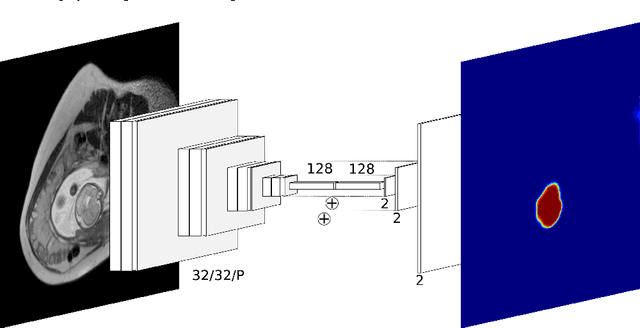
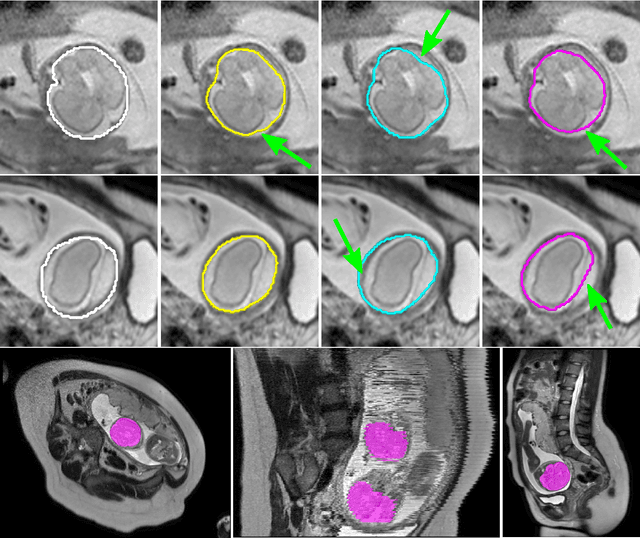
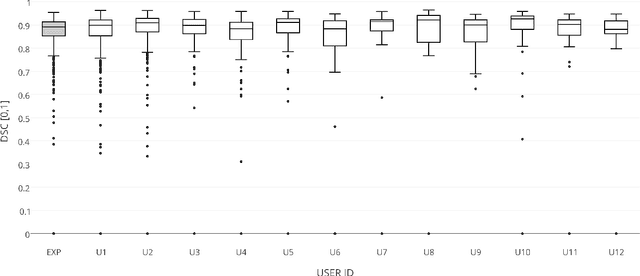
Abstract:The availability of training data for supervision is a frequently encountered bottleneck of medical image analysis methods. While typically established by a clinical expert rater, the increase in acquired imaging data renders traditional pixel-wise segmentations less feasible. In this paper, we examine the use of a crowdsourcing platform for the distribution of super-pixel weak annotation tasks and collect such annotations from a crowd of non-expert raters. The crowd annotations are subsequently used for training a fully convolutional neural network to address the problem of fetal brain segmentation in T2-weighted MR images. Using this approach we report encouraging results compared to highly targeted, fully supervised methods and potentially address a frequent problem impeding image analysis research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge