Denis Prokopenko
DCRA-Net: Attention-Enabled Reconstruction Model for Dynamic Fetal Cardiac MRI
Dec 19, 2024



Abstract:Dynamic fetal heart magnetic resonance imaging (MRI) presents unique challenges due to the fast heart rate of the fetus compared to adult subjects and uncontrolled fetal motion. This requires high temporal and spatial resolutions over a large field of view, in order to encompass surrounding maternal anatomy. In this work, we introduce Dynamic Cardiac Reconstruction Attention Network (DCRA-Net) - a novel deep learning model that employs attention mechanisms in spatial and temporal domains and temporal frequency representation of data to reconstruct the dynamics of the fetal heart from highly accelerated free-running (non-gated) MRI acquisitions. DCRA-Net was trained on retrospectively undersampled complex-valued cardiac MRIs from 42 fetal subjects and separately from 153 adult subjects, and evaluated on data from 14 fetal and 39 adult subjects respectively. Its performance was compared to L+S and k-GIN methods in both fetal and adult cases for an undersampling factor of 8x. The proposed network performed better than the comparators for both fetal and adult data, for both regular lattice and centrally weighted random undersampling. Aliased signals due to the undersampling were comprehensively resolved, and both the spatial details of the heart and its temporal dynamics were recovered with high fidelity. The highest performance was achieved when using lattice undersampling, data consistency and temporal frequency representation, yielding PSNR of 38 for fetal and 35 for adult cases. Our method is publicly available at https://github.com/denproc/DCRA-Net.
QUASAR: QUality and Aesthetics Scoring with Advanced Representations
Mar 20, 2024



Abstract:This paper introduces a new data-driven, non-parametric method for image quality and aesthetics assessment, surpassing existing approaches and requiring no prompt engineering or fine-tuning. We eliminate the need for expressive textual embeddings by proposing efficient image anchors in the data. Through extensive evaluations of 7 state-of-the-art self-supervised models, our method demonstrates superior performance and robustness across various datasets and benchmarks. Notably, it achieves high agreement with human assessments even with limited data and shows high robustness to the nature of data and their pre-processing pipeline. Our contributions offer a streamlined solution for assessment of images while providing insights into the perception of visual information.
The Challenge of Fetal Cardiac MRI Reconstruction Using Deep Learning
Aug 15, 2023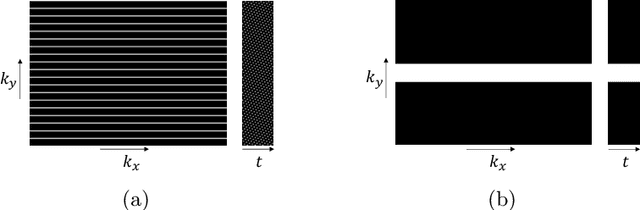
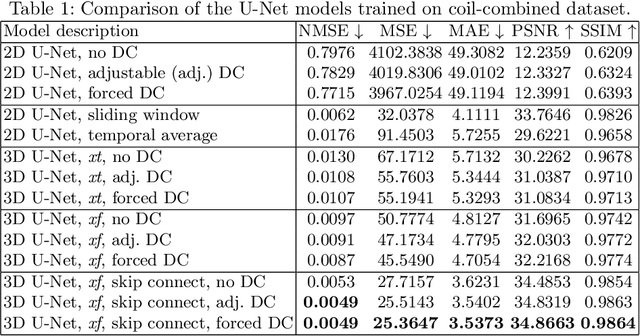
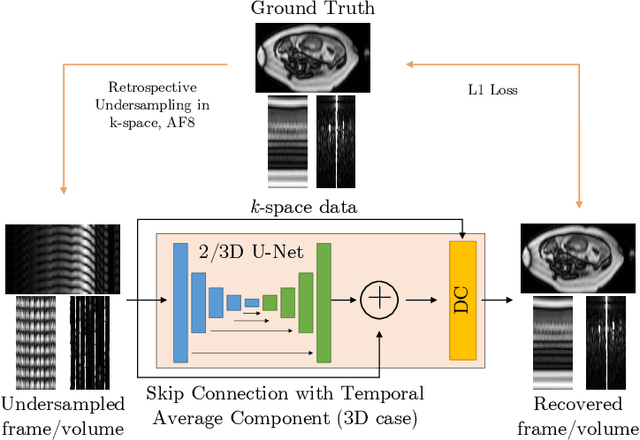
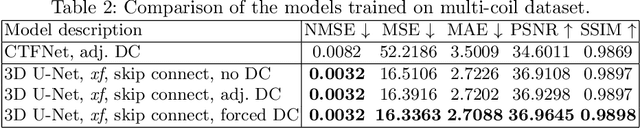
Abstract:Dynamic free-breathing fetal cardiac MRI is one of the most challenging modalities, which requires high temporal and spatial resolution to depict rapid changes in a small fetal heart. The ability of deep learning methods to recover undersampled data could help to optimise the kt-SENSE acquisition strategy and improve non-gated kt-SENSE reconstruction quality. In this work, we explore supervised deep learning networks for reconstruction of kt-SENSE style acquired data using an extensive in vivo dataset. Having access to fully-sampled low-resolution multi-coil fetal cardiac MRI, we study the performance of the networks to recover fully-sampled data from undersampled data. We consider model architectures together with training strategies taking into account their application in the real clinical setup used to collect the dataset to enable networks to recover prospectively undersampled data. We explore a set of modifications to form a baseline performance evaluation for dynamic fetal cardiac MRI on real data. We systematically evaluate the models on coil-combined data to reveal the effect of the suggested changes to the architecture in the context of fetal heart properties. We show that the best-performers recover a detailed depiction of the maternal anatomy on a large scale, but the dynamic properties of the fetal heart are under-represented. Training directly on multi-coil data improves the performance of the models, allows their prospective application to undersampled data and makes them outperform CTFNet introduced for adult cardiac cine MRI. However, these models deliver similar qualitative performances recovering the maternal body very well but underestimating the dynamic properties of fetal heart. This dynamic feature of fast change of fetal heart that is highly localised suggests both more targeted training and evaluation methods might be needed for fetal heart application.
PyTorch Image Quality: Metrics for Image Quality Assessment
Aug 31, 2022
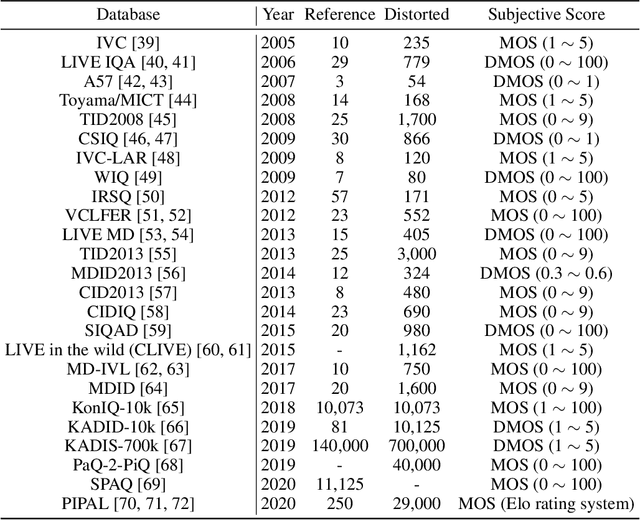
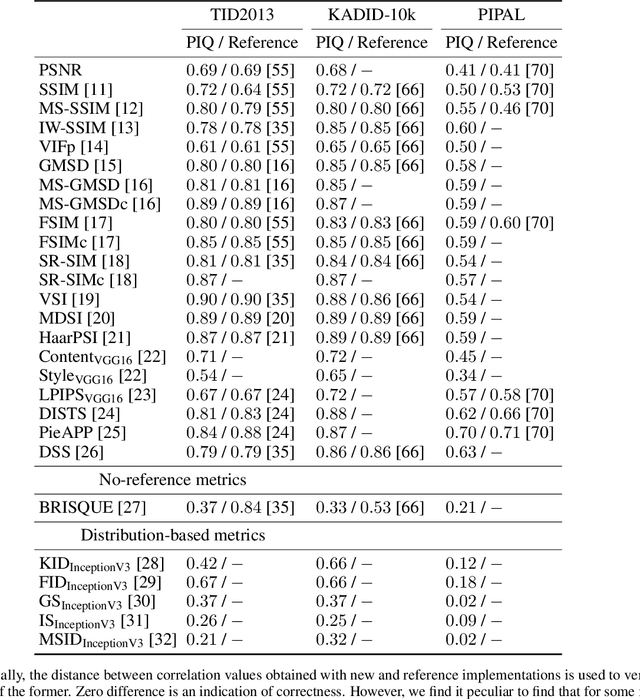
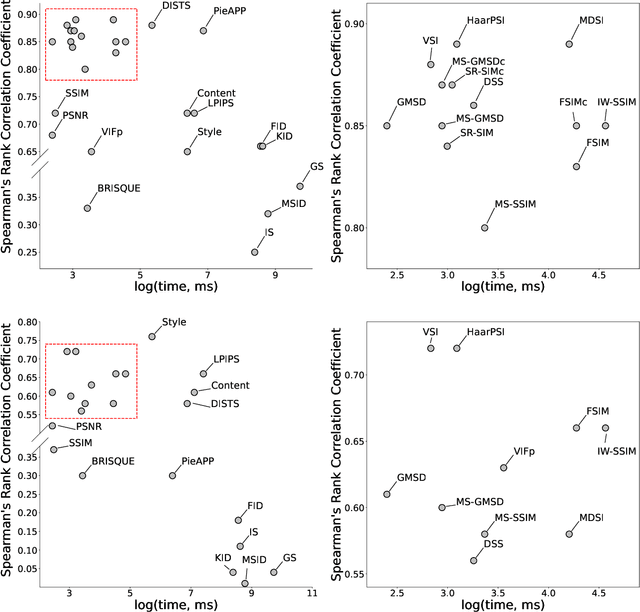
Abstract:Image Quality Assessment (IQA) metrics are widely used to quantitatively estimate the extent of image degradation following some forming, restoring, transforming, or enhancing algorithms. We present PyTorch Image Quality (PIQ), a usability-centric library that contains the most popular modern IQA algorithms, guaranteed to be correctly implemented according to their original propositions and thoroughly verified. In this paper, we detail the principles behind the foundation of the library, describe the evaluation strategy that makes it reliable, provide the benchmarks that showcase the performance-time trade-offs, and underline the benefits of GPU acceleration given the library is used within the PyTorch backend. PyTorch Image Quality is an open source software: https://github.com/photosynthesis-team/piq/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge