Philippe Lambin
on behalf of EUCanImage working group
Robust Multicentre Detection and Classification of Colorectal Liver Metastases on CT: Application of Foundation Models
Jan 12, 2026Abstract:Colorectal liver metastases (CRLM) are a major cause of cancer-related mortality, and reliable detection on CT remains challenging in multi-centre settings. We developed a foundation model-based AI pipeline for patient-level classification and lesion-level detection of CRLM on contrast-enhanced CT, integrating uncertainty quantification and explainability. CT data from the EuCanImage consortium (n=2437) and an external TCIA cohort (n=197) were used. Among several pretrained models, UMedPT achieved the best performance and was fine-tuned with an MLP head for classification and an FCOS-based head for lesion detection. The classification model achieved an AUC of 0.90 and a sensitivity of 0.82 on the combined test set, with a sensitivity of 0.85 on the external cohort. Excluding the most uncertain 20 percent of cases improved AUC to 0.91 and balanced accuracy to 0.86. Decision curve analysis showed clinical benefit for threshold probabilities between 0.30 and 0.40. The detection model identified 69.1 percent of lesions overall, increasing from 30 percent to 98 percent across lesion size quartiles. Grad-CAM highlighted lesion-corresponding regions in high-confidence cases. These results demonstrate that foundation model-based pipelines can support robust and interpretable CRLM detection and classification across heterogeneous CT data.
Radiology Report Conditional 3D CT Generation with Multi Encoder Latent diffusion Model
Sep 18, 2025



Abstract:Text to image latent diffusion models have recently advanced medical image synthesis, but applications to 3D CT generation remain limited. Existing approaches rely on simplified prompts, neglecting the rich semantic detail in full radiology reports, which reduces text image alignment and clinical fidelity. We propose Report2CT, a radiology report conditional latent diffusion framework for synthesizing 3D chest CT volumes directly from free text radiology reports, incorporating both findings and impression sections using multiple text encoder. Report2CT integrates three pretrained medical text encoders (BiomedVLP CXR BERT, MedEmbed, and ClinicalBERT) to capture nuanced clinical context. Radiology reports and voxel spacing information condition a 3D latent diffusion model trained on 20000 CT volumes from the CT RATE dataset. Model performance was evaluated using Frechet Inception Distance (FID) for real synthetic distributional similarity and CLIP based metrics for semantic alignment, with additional qualitative and quantitative comparisons against GenerateCT model. Report2CT generated anatomically consistent CT volumes with excellent visual quality and text image alignment. Multi encoder conditioning improved CLIP scores, indicating stronger preservation of fine grained clinical details in the free text radiology reports. Classifier free guidance further enhanced alignment with only a minor trade off in FID. We ranked first in the VLM3D Challenge at MICCAI 2025 on Text Conditional CT Generation and achieved state of the art performance across all evaluation metrics. By leveraging complete radiology reports and multi encoder text conditioning, Report2CT advances 3D CT synthesis, producing clinically faithful and high quality synthetic data.
Explainable Anatomy-Guided AI for Prostate MRI: Foundation Models and In Silico Clinical Trials for Virtual Biopsy-based Risk Assessment
May 23, 2025Abstract:We present a fully automated, anatomically guided deep learning pipeline for prostate cancer (PCa) risk stratification using routine MRI. The pipeline integrates three key components: an nnU-Net module for segmenting the prostate gland and its zones on axial T2-weighted MRI; a classification module based on the UMedPT Swin Transformer foundation model, fine-tuned on 3D patches with optional anatomical priors and clinical data; and a VAE-GAN framework for generating counterfactual heatmaps that localize decision-driving image regions. The system was developed using 1,500 PI-CAI cases for segmentation and 617 biparametric MRIs with metadata from the CHAIMELEON challenge for classification (split into 70% training, 10% validation, and 20% testing). Segmentation achieved mean Dice scores of 0.95 (gland), 0.94 (peripheral zone), and 0.92 (transition zone). Incorporating gland priors improved AUC from 0.69 to 0.72, with a three-scale ensemble achieving top performance (AUC = 0.79, composite score = 0.76), outperforming the 2024 CHAIMELEON challenge winners. Counterfactual heatmaps reliably highlighted lesions within segmented regions, enhancing model interpretability. In a prospective multi-center in-silico trial with 20 clinicians, AI assistance increased diagnostic accuracy from 0.72 to 0.77 and Cohen's kappa from 0.43 to 0.53, while reducing review time per case by 40%. These results demonstrate that anatomy-aware foundation models with counterfactual explainability can enable accurate, interpretable, and efficient PCa risk assessment, supporting their potential use as virtual biopsies in clinical practice.
Pixels to Prognosis: Harmonized Multi-Region CT-Radiomics and Foundation-Model Signatures Across Multicentre NSCLC Data
May 23, 2025Abstract:Purpose: To evaluate the impact of harmonization and multi-region CT image feature integration on survival prediction in non-small cell lung cancer (NSCLC) patients, using handcrafted radiomics, pretrained foundation model (FM) features, and clinical data from a multicenter dataset. Methods: We analyzed CT scans and clinical data from 876 NSCLC patients (604 training, 272 test) across five centers. Features were extracted from the whole lung, tumor, mediastinal nodes, coronary arteries, and coronary artery calcium (CAC). Handcrafted radiomics and FM deep features were harmonized using ComBat, reconstruction kernel normalization (RKN), and RKN+ComBat. Regularized Cox models predicted overall survival; performance was assessed using the concordance index (C-index), 5-year time-dependent area under the curve (t-AUC), and hazard ratio (HR). SHapley Additive exPlanations (SHAP) values explained feature contributions. A consensus model used agreement across top region of interest (ROI) models to stratify patient risk. Results: TNM staging showed prognostic utility (C-index = 0.67; HR = 2.70; t-AUC = 0.85). The clinical + tumor radiomics model with ComBat achieved a C-index of 0.7552 and t-AUC of 0.8820. FM features (50-voxel cubes) combined with clinical data yielded the highest performance (C-index = 0.7616; t-AUC = 0.8866). An ensemble of all ROIs and FM features reached a C-index of 0.7142 and t-AUC of 0.7885. The consensus model, covering 78% of valid test cases, achieved a t-AUC of 0.92, sensitivity of 97.6%, and specificity of 66.7%. Conclusion: Harmonization and multi-region feature integration improve survival prediction in multicenter NSCLC data. Combining interpretable radiomics, FM features, and consensus modeling enables robust risk stratification across imaging centers.
A Foundation Model Framework for Multi-View MRI Classification of Extramural Vascular Invasion and Mesorectal Fascia Invasion in Rectal Cancer
May 23, 2025Abstract:Background: Accurate MRI-based identification of extramural vascular invasion (EVI) and mesorectal fascia invasion (MFI) is pivotal for risk-stratified management of rectal cancer, yet visual assessment is subjective and vulnerable to inter-institutional variability. Purpose: To develop and externally evaluate a multicenter, foundation-model-driven framework that automatically classifies EVI and MFI on axial and sagittal T2-weighted MRI. Methods: This retrospective study used 331 pre-treatment rectal cancer MRI examinations from three European hospitals. After TotalSegmentator-guided rectal patch extraction, a self-supervised frequency-domain harmonization pipeline was trained to minimize scanner-related contrast shifts. Four classifiers were compared: ResNet50, SeResNet, the universal biomedical pretrained transformer (UMedPT) with a lightweight MLP head, and a logistic-regression variant using frozen UMedPT features (UMedPT_LR). Results: UMedPT_LR achieved the best EVI detection when axial and sagittal features were fused (AUC = 0.82; sensitivity = 0.75; F1 score = 0.73), surpassing the Chaimeleon Grand-Challenge winner (AUC = 0.74). The highest MFI performance was attained by UMedPT on axial harmonized images (AUC = 0.77), surpassing the Chaimeleon Grand-Challenge winner (AUC = 0.75). Frequency-domain harmonization improved MFI classification but variably affected EVI performance. Conventional CNNs (ResNet50, SeResNet) underperformed, especially in F1 score and balanced accuracy. Conclusion: These findings demonstrate that combining foundation model features, harmonization, and multi-view fusion significantly enhances diagnostic performance in rectal MRI.
Advancing oncology with federated learning: transcending boundaries in breast, lung, and prostate cancer. A systematic review
Aug 08, 2024Abstract:Federated Learning (FL) has emerged as a promising solution to address the limitations of centralised machine learning (ML) in oncology, particularly in overcoming privacy concerns and harnessing the power of diverse, multi-center data. This systematic review synthesises current knowledge on the state-of-the-art FL in oncology, focusing on breast, lung, and prostate cancer. Distinct from previous surveys, our comprehensive review critically evaluates the real-world implementation and impact of FL on cancer care, demonstrating its effectiveness in enhancing ML generalisability, performance and data privacy in clinical settings and data. We evaluated state-of-the-art advances in FL, demonstrating its growing adoption amid tightening data privacy regulations. FL outperformed centralised ML in 15 out of the 25 studies reviewed, spanning diverse ML models and clinical applications, and facilitating integration of multi-modal information for precision medicine. Despite the current challenges identified in reproducibility, standardisation and methodology across studies, the demonstrable benefits of FL in harnessing real-world data and addressing clinical needs highlight its significant potential for advancing cancer research. We propose that future research should focus on addressing these limitations and investigating further advanced FL methods, to fully harness data diversity and realise the transformative power of cutting-edge FL in cancer care.
Counterfactuals and Uncertainty-Based Explainable Paradigm for the Automated Detection and Segmentation of Renal Cysts in Computed Tomography Images: A Multi-Center Study
Aug 07, 2024Abstract:Routine computed tomography (CT) scans often detect a wide range of renal cysts, some of which may be malignant. Early and precise localization of these cysts can significantly aid quantitative image analysis. Current segmentation methods, however, do not offer sufficient interpretability at the feature and pixel levels, emphasizing the necessity for an explainable framework that can detect and rectify model inaccuracies. We developed an interpretable segmentation framework and validated it on a multi-centric dataset. A Variational Autoencoder Generative Adversarial Network (VAE-GAN) was employed to learn the latent representation of 3D input patches and reconstruct input images. Modifications in the latent representation using the gradient of the segmentation model generated counterfactual explanations for varying dice similarity coefficients (DSC). Radiomics features extracted from these counterfactual images, using a ground truth cyst mask, were analyzed to determine their correlation with segmentation performance. The DSCs for the original and VAE-GAN reconstructed images for counterfactual image generation showed no significant differences. Counterfactual explanations highlighted how variations in cyst image features influence segmentation outcomes and showed model discrepancies. Radiomics features correlating positively and negatively with dice scores were identified. The uncertainty of the predicted segmentation masks was estimated using posterior sampling of the weight space. The combination of counterfactual explanations and uncertainty maps provided a deeper understanding of the image features within the segmented renal cysts that lead to high uncertainty. The proposed segmentation framework not only achieved high segmentation accuracy but also increased interpretability regarding how image features impact segmentation performance.
Methodological Explainability Evaluation of an Interpretable Deep Learning Model for Post-Hepatectomy Liver Failure Prediction Incorporating Counterfactual Explanations and Layerwise Relevance Propagation: A Prospective In Silico Trial
Aug 07, 2024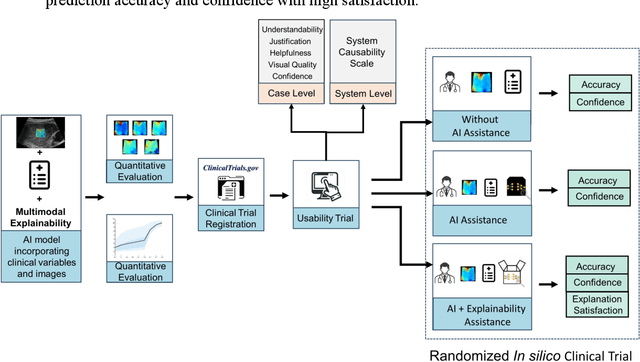
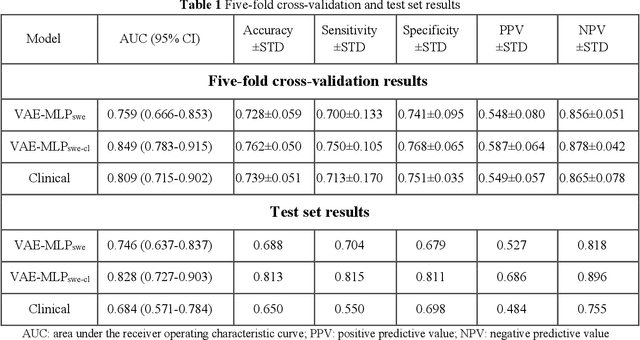
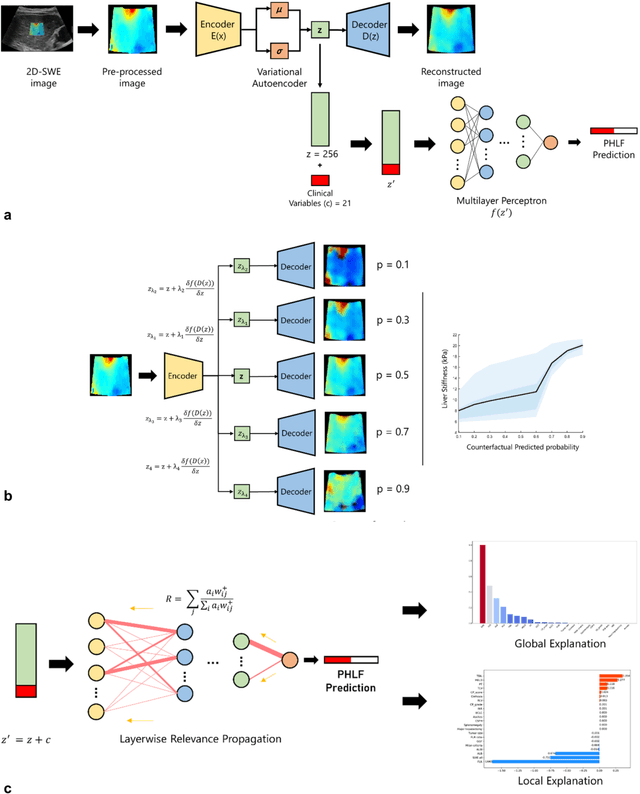
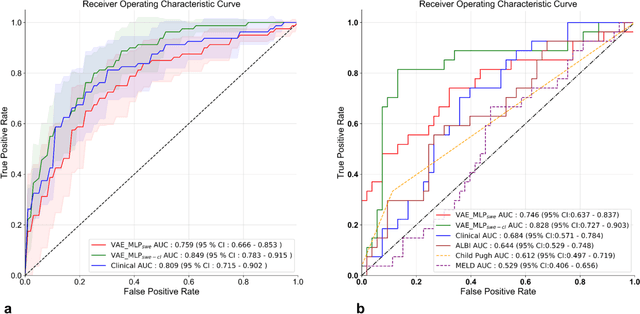
Abstract:Artificial intelligence (AI)-based decision support systems have demonstrated value in predicting post-hepatectomy liver failure (PHLF) in hepatocellular carcinoma (HCC). However, they often lack transparency, and the impact of model explanations on clinicians' decisions has not been thoroughly evaluated. Building on prior research, we developed a variational autoencoder-multilayer perceptron (VAE-MLP) model for preoperative PHLF prediction. This model integrated counterfactuals and layerwise relevance propagation (LRP) to provide insights into its decision-making mechanism. Additionally, we proposed a methodological framework for evaluating the explainability of AI systems. This framework includes qualitative and quantitative assessments of explanations against recognized biomarkers, usability evaluations, and an in silico clinical trial. Our evaluations demonstrated that the model's explanation correlated with established biomarkers and exhibited high usability at both the case and system levels. Furthermore, results from the three-track in silico clinical trial showed that clinicians' prediction accuracy and confidence increased when AI explanations were provided.
A review of handcrafted and deep radiomics in neurological diseases: transitioning from oncology to clinical neuroimaging
Jul 18, 2024



Abstract:Medical imaging technologies have undergone extensive development, enabling non-invasive visualization of clinical information. The traditional review of medical images by clinicians remains subjective, time-consuming, and prone to human error. With the recent availability of medical imaging data, quantification have become important goals in the field. Radiomics, a methodology aimed at extracting quantitative information from imaging data, has emerged as a promising approach to uncover hidden biological information and support decision-making in clinical practice. This paper presents a review of the radiomic pipeline from the clinical neuroimaging perspective, providing a detailed overview of each step with practical advice. It discusses the application of handcrafted and deep radiomics in neuroimaging, stratified by neurological diagnosis. Although radiomics shows great potential for increasing diagnostic precision and improving treatment quality in neurology, several limitations hinder its clinical implementation. Addressing these challenges requires collaborative efforts, advancements in image harmonization methods, and the establishment of reproducible and standardized pipelines with transparent reporting. By overcoming these obstacles, radiomics can significantly impact clinical neurology and enhance patient care.
MSCDA: Multi-level Semantic-guided Contrast Improves Unsupervised Domain Adaptation for Breast MRI Segmentation in Small Datasets
Jan 04, 2023



Abstract:Deep learning (DL) applied to breast tissue segmentation in magnetic resonance imaging (MRI) has received increased attention in the last decade, however, the domain shift which arises from different vendors, acquisition protocols, and biological heterogeneity, remains an important but challenging obstacle on the path towards clinical implementation. Recently, unsupervised domain adaptation (UDA) methods have attempted to mitigate this problem by incorporating self-training with contrastive learning. To better exploit the underlying semantic information of the image at different levels, we propose a Multi-level Semantic-guided Contrastive Domain Adaptation (MSCDA) framework to align the feature representation between domains. In particular, we extend the contrastive loss by incorporating pixel-to-pixel, pixel-to-centroid, and centroid-to-centroid contrasts to integrate semantic information of images. We utilize a category-wise cross-domain sampling strategy to sample anchors from target images and build a hybrid memory bank to store samples from source images. Two breast MRI datasets were retrospectively collected: The source dataset contains non-contrast MRI examinations from 11 healthy volunteers and the target dataset contains contrast-enhanced MRI examinations of 134 invasive breast cancer patients. We set up experiments from source T2W image to target dynamic contrast-enhanced (DCE)-T1W image (T2W-to-T1W) and from source T1W image to target T2W image (T1W-to-T2W). The proposed method achieved Dice similarity coefficient (DSC) of 89.2\% and 84.0\% in T2W-to-T1W and T1W-to-T2W, respectively, outperforming state-of-the-art methods. Notably, good performance is still achieved with a smaller source dataset, proving that our framework is label-efficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge