Michael Fitzke
VET-DINO: Learning Anatomical Understanding Through Multi-View Distillation in Veterinary Imaging
May 21, 2025Abstract:Self-supervised learning has emerged as a powerful paradigm for training deep neural networks, particularly in medical imaging where labeled data is scarce. While current approaches typically rely on synthetic augmentations of single images, we propose VET-DINO, a framework that leverages a unique characteristic of medical imaging: the availability of multiple standardized views from the same study. Using a series of clinical veterinary radiographs from the same patient study, we enable models to learn view-invariant anatomical structures and develop an implied 3D understanding from 2D projections. We demonstrate our approach on a dataset of 5 million veterinary radiographs from 668,000 canine studies. Through extensive experimentation, including view synthesis and downstream task performance, we show that learning from real multi-view pairs leads to superior anatomical understanding compared to purely synthetic augmentations. VET-DINO achieves state-of-the-art performance on various veterinary imaging tasks. Our work establishes a new paradigm for self-supervised learning in medical imaging that leverages domain-specific properties rather than merely adapting natural image techniques.
PulseNet: Deep Learning ECG-signal classification using random augmentation policy and continous wavelet transform for canines
May 17, 2023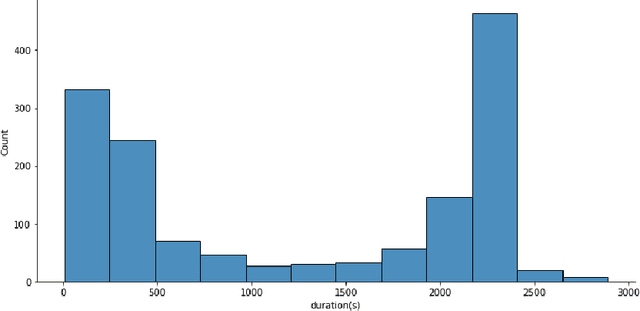
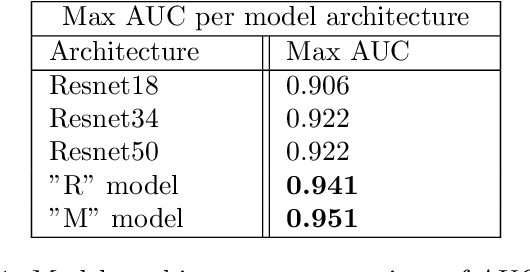
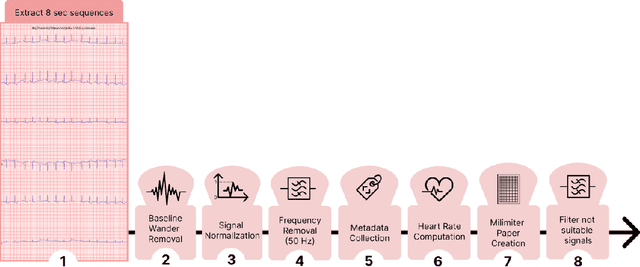
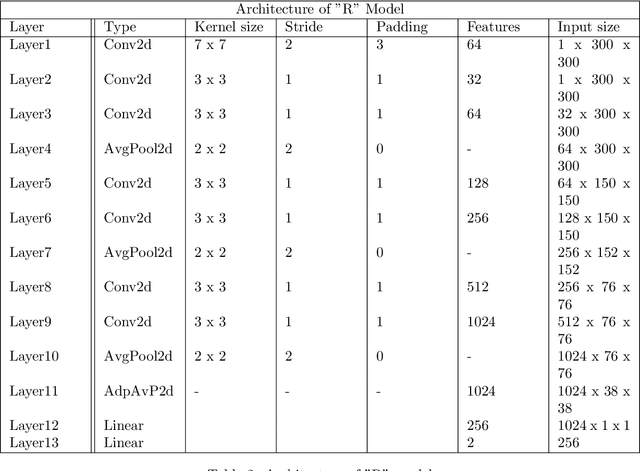
Abstract:Evaluating canine electrocardiograms (ECG) require skilled veterinarians, but current availability of veterinary cardiologists for ECG interpretation and diagnostic support is limited. Developing tools for automated assessment of ECG sequences can improve veterinary care by providing clinicians real-time results and decision support tools. We implement a deep convolutional neural network (CNN) approach for classifying canine electrocardiogram sequences as either normal or abnormal. ECG records are converted into 8 second Lead II sequences and classified as either normal (no evidence of cardiac abnormalities) or abnormal (presence of one or more cardiac abnormalities). For training ECG sequences are randomly augmented using RandomAugmentECG, a new augmentation library implemented specifically for this project. Each chunk is then is converted using a continuous wavelet transform into a 2D scalogram. The 2D scalogram are then classified as either normal or abnormal by a binary CNN classifier. Experimental results are validated against three boarded veterinary cardiologists achieving an AUC-ROC score of 0.9506 on test dataset matching human level performance. Additionally, we describe model deployment to Microsoft Azure using an MLOps approach. To our knowledge, this work is one of the first attempts to implement a deep learning model to automatically classify ECG sequences for canines.Implementing automated ECG classification will enhance veterinary care through improved diagnostic performance and increased clinic efficiency.
StudyFormer : Attention-Based and Dynamic Multi View Classifier for X-ray images
Feb 23, 2023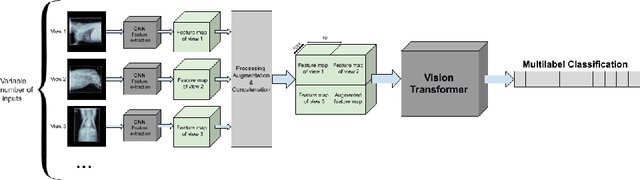
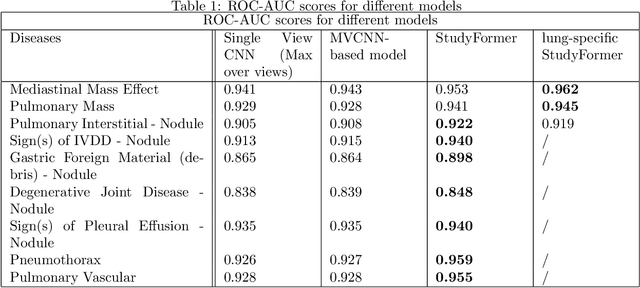
Abstract:Chest X-ray images are commonly used in medical diagnosis, and AI models have been developed to assist with the interpretation of these images. However, many of these models rely on information from a single view of the X-ray, while multiple views may be available. In this work, we propose a novel approach for combining information from multiple views to improve the performance of X-ray image classification. Our approach is based on the use of a convolutional neural network to extract feature maps from each view, followed by an attention mechanism implemented using a Vision Transformer. The resulting model is able to perform multi-label classification on 41 labels and outperforms both single-view models and traditional multi-view classification architectures. We demonstrate the effectiveness of our approach through experiments on a dataset of 363,000 X-ray images.
Who Goes First? Influences of Human-AI Workflow on Decision Making in Clinical Imaging
May 19, 2022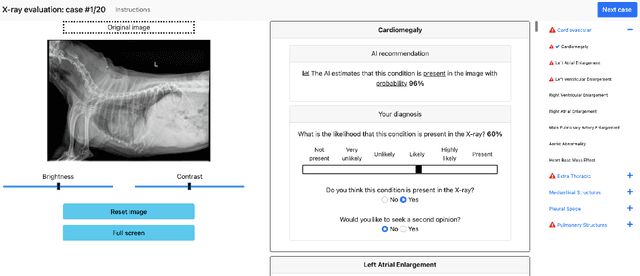
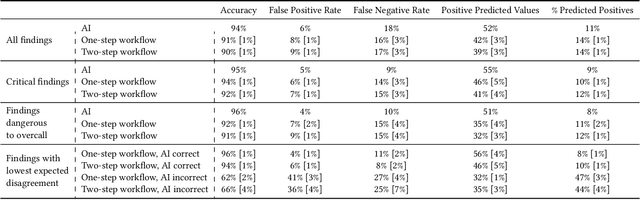
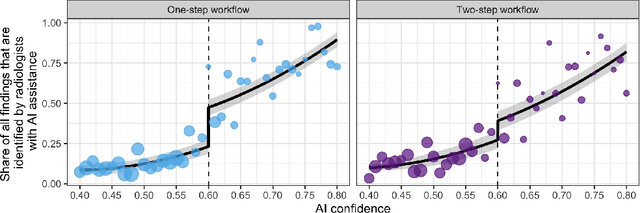
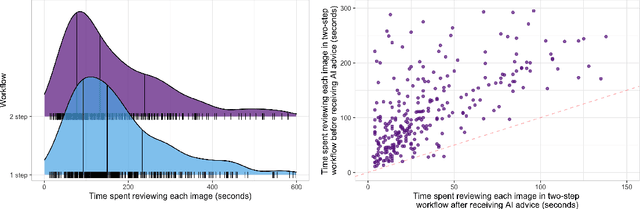
Abstract:Details of the designs and mechanisms in support of human-AI collaboration must be considered in the real-world fielding of AI technologies. A critical aspect of interaction design for AI-assisted human decision making are policies about the display and sequencing of AI inferences within larger decision-making workflows. We have a poor understanding of the influences of making AI inferences available before versus after human review of a diagnostic task at hand. We explore the effects of providing AI assistance at the start of a diagnostic session in radiology versus after the radiologist has made a provisional decision. We conducted a user study where 19 veterinary radiologists identified radiographic findings present in patients' X-ray images, with the aid of an AI tool. We employed two workflow configurations to analyze (i) anchoring effects, (ii) human-AI team diagnostic performance and agreement, (iii) time spent and confidence in decision making, and (iv) perceived usefulness of the AI. We found that participants who are asked to register provisional responses in advance of reviewing AI inferences are less likely to agree with the AI regardless of whether the advice is accurate and, in instances of disagreement with the AI, are less likely to seek the second opinion of a colleague. These participants also reported the AI advice to be less useful. Surprisingly, requiring provisional decisions on cases in advance of the display of AI inferences did not lengthen the time participants spent on the task. The study provides generalizable and actionable insights for the deployment of clinical AI tools in human-in-the-loop systems and introduces a methodology for studying alternative designs for human-AI collaboration. We make our experimental platform available as open source to facilitate future research on the influence of alternate designs on human-AI workflows.
RapidRead: Global Deployment of State-of-the-art Radiology AI for a Large Veterinary Teleradiology Practice
Nov 09, 2021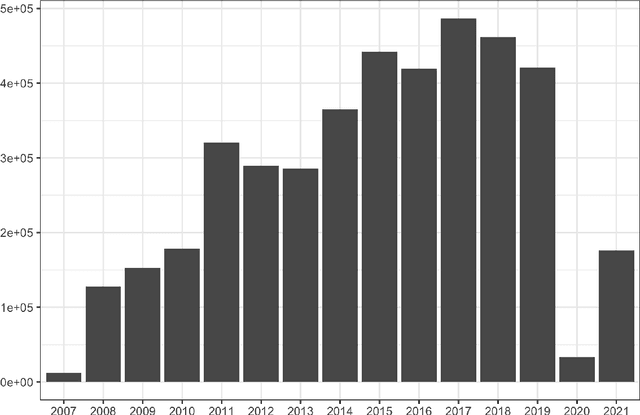
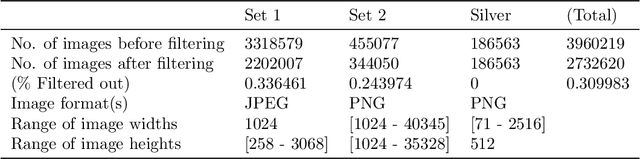
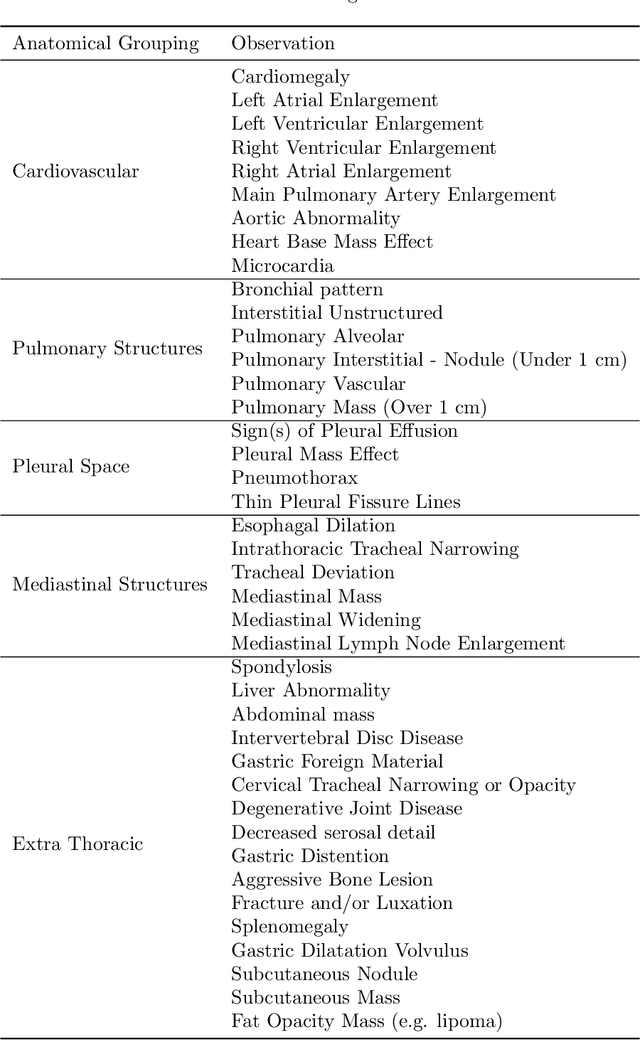
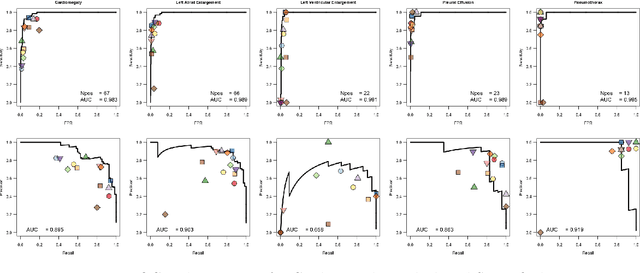
Abstract:This work describes the development and real-world deployment of a deep learning-based AI system for evaluating canine and feline radiographs across a broad range of findings and abnormalities. We describe a new semi-supervised learning approach that combines NLP-derived labels with self-supervised training leveraging more than 2.5 million x-ray images. Finally we describe the clinical deployment of the model including system architecture, real-time performance evaluation and data drift detection.
OncoPetNet: A Deep Learning based AI system for mitotic figure counting on H&E stained whole slide digital images in a large veterinary diagnostic lab setting
Aug 17, 2021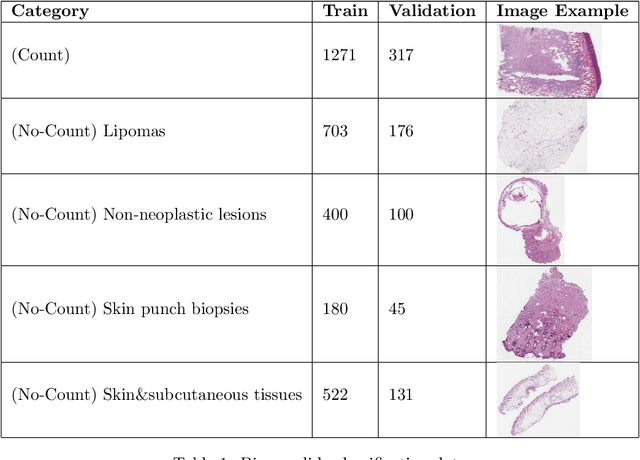
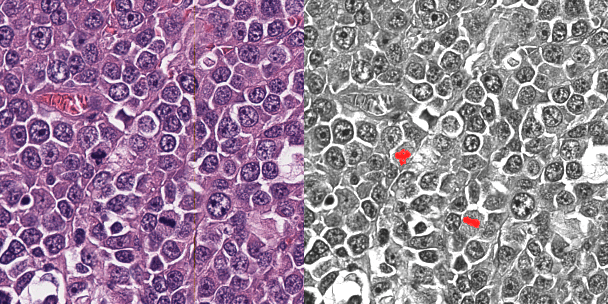
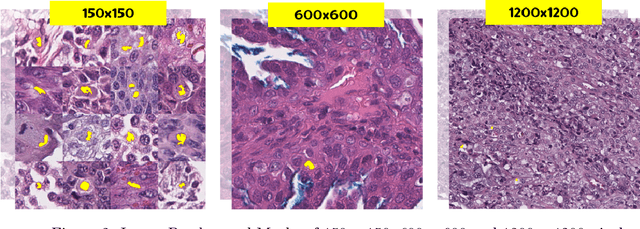
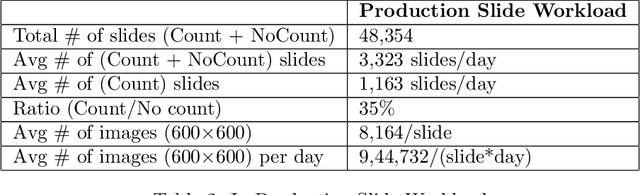
Abstract:Background: Histopathology is an important modality for the diagnosis and management of many diseases in modern healthcare, and plays a critical role in cancer care. Pathology samples can be large and require multi-site sampling, leading to upwards of 20 slides for a single tumor, and the human-expert tasks of site selection and and quantitative assessment of mitotic figures are time consuming and subjective. Automating these tasks in the setting of a digital pathology service presents significant opportunities to improve workflow efficiency and augment human experts in practice. Approach: Multiple state-of-the-art deep learning techniques for histopathology image classification and mitotic figure detection were used in the development of OncoPetNet. Additionally, model-free approaches were used to increase speed and accuracy. The robust and scalable inference engine leverages Pytorch's performance optimizations as well as specifically developed speed up techniques in inference. Results: The proposed system, demonstrated significantly improved mitotic counting performance for 41 cancer cases across 14 cancer types compared to human expert baselines. In 21.9% of cases use of OncoPetNet led to change in tumor grading compared to human expert evaluation. In deployment, an effective 0.27 min/slide inference was achieved in a high throughput veterinary diagnostic pathology service across 2 centers processing 3,323 digital whole slide images daily. Conclusion: This work represents the first successful automated deployment of deep learning systems for real-time expert-level performance on important histopathology tasks at scale in a high volume clinical practice. The resulting impact outlines important considerations for model development, deployment, clinical decision making, and informs best practices for implementation of deep learning systems in digital histopathology practices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge