Chris Carter
OncoPetNet: A Deep Learning based AI system for mitotic figure counting on H&E stained whole slide digital images in a large veterinary diagnostic lab setting
Aug 17, 2021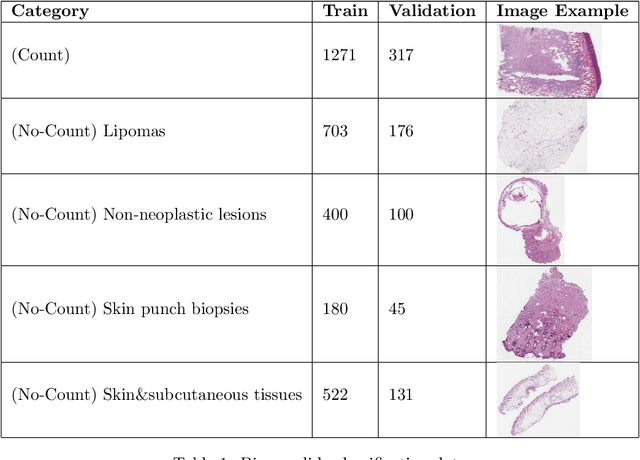
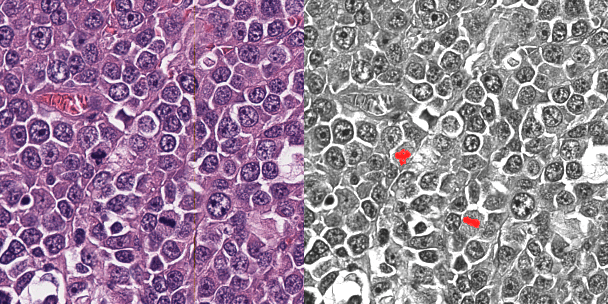
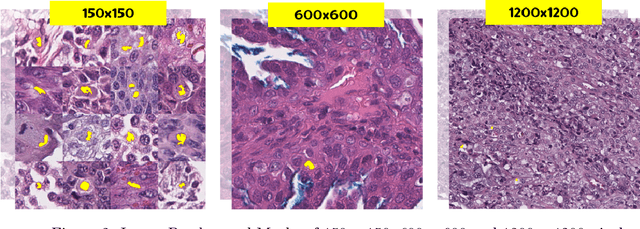
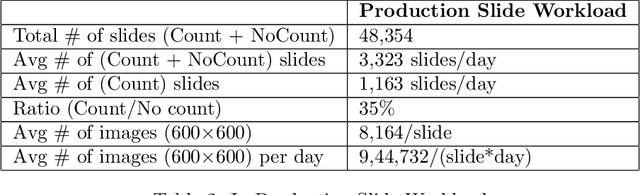
Abstract:Background: Histopathology is an important modality for the diagnosis and management of many diseases in modern healthcare, and plays a critical role in cancer care. Pathology samples can be large and require multi-site sampling, leading to upwards of 20 slides for a single tumor, and the human-expert tasks of site selection and and quantitative assessment of mitotic figures are time consuming and subjective. Automating these tasks in the setting of a digital pathology service presents significant opportunities to improve workflow efficiency and augment human experts in practice. Approach: Multiple state-of-the-art deep learning techniques for histopathology image classification and mitotic figure detection were used in the development of OncoPetNet. Additionally, model-free approaches were used to increase speed and accuracy. The robust and scalable inference engine leverages Pytorch's performance optimizations as well as specifically developed speed up techniques in inference. Results: The proposed system, demonstrated significantly improved mitotic counting performance for 41 cancer cases across 14 cancer types compared to human expert baselines. In 21.9% of cases use of OncoPetNet led to change in tumor grading compared to human expert evaluation. In deployment, an effective 0.27 min/slide inference was achieved in a high throughput veterinary diagnostic pathology service across 2 centers processing 3,323 digital whole slide images daily. Conclusion: This work represents the first successful automated deployment of deep learning systems for real-time expert-level performance on important histopathology tasks at scale in a high volume clinical practice. The resulting impact outlines important considerations for model development, deployment, clinical decision making, and informs best practices for implementation of deep learning systems in digital histopathology practices.
Multiple decision trees
Mar 27, 2013


Abstract:This paper describes experiments, on two domains, to investigate the effect of averaging over predictions of multiple decision trees, instead of using a single tree. Other authors have pointed out theoretical and commonsense reasons for preferring the multiple tree approach. Ideally, we would like to consider predictions from all trees, weighted by their probability. However, there is a vast number of different trees, and it is difficult to estimate the probability of each tree. We sidestep the estimation problem by using a modified version of the ID3 algorithm to build good trees, and average over only these trees. Our results are encouraging. For each domain, we managed to produce a small number of good trees. We find that it is best to average across sets of trees with different structure; this usually gives better performance than any of the constituent trees, including the ID3 tree.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge