Evan Zucker
Scout-Net: Prospective Personalized Estimation of CT Organ Doses from Scout Views
Dec 23, 2023Abstract:Purpose: Estimation of patient-specific organ doses is required for more comprehensive dose metrics, such as effective dose. Currently, available methods are performed retrospectively using the CT images themselves, which can only be done after the scan. To optimize CT acquisitions before scanning, rapid prediction of patient-specific organ dose is needed prospectively, using available scout images. We, therefore, devise an end-to-end, fully-automated deep learning solution to perform real-time, patient-specific, organ-level dosimetric estimation of CT scans. Approach: We propose the Scout-Net model for CT dose prediction at six different organs as well as for the overall patient body, leveraging the routinely obtained frontal and lateral scout images of patients, before their CT scans. To obtain reference values of the organ doses, we used Monte Carlo simulation and 3D segmentation methods on the corresponding CT images of the patients. Results: We validate our proposed Scout-Net model against real patient CT data and demonstrate the effectiveness in estimating organ doses in real-time (only 27 ms on average per scan). Additionally, we demonstrate the efficiency (real-time execution), sufficiency (reasonable error rates), and robustness (consistent across varying patient sizes) of the Scout-Net model. Conclusions: An effective, efficient, and robust Scout-Net model, once incorporated into the CT acquisition plan, could potentially guide the automatic exposure control for balanced image quality and radiation dose.
The Effect of Counterfactuals on Reading Chest X-rays
Apr 02, 2023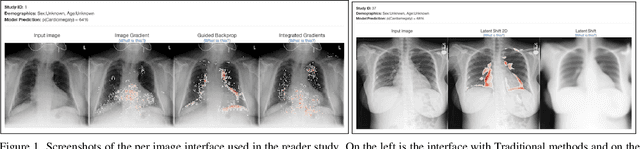
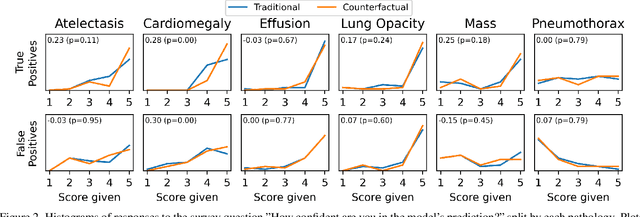
Abstract:This study evaluates the effect of counterfactual explanations on the interpretation of chest X-rays. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to rate their confidence that the model's prediction is correct using a 5 point scale. Half of the predictions are false positives. Each prediction is explained twice, once using traditional attribution methods and once with a counterfactual explanation. The overall results indicate that counterfactual explanations allow a radiologist to have more confidence in true positive predictions compared to traditional approaches (0.15$\pm$0.95 with p=0.01) with only a small increase in false positive predictions (0.04$\pm$1.06 with p=0.57). We observe the specific prediction tasks of Mass and Atelectasis appear to benefit the most compared to other tasks.
Gifsplanation via Latent Shift: A Simple Autoencoder Approach to Progressive Exaggeration on Chest X-rays
Feb 18, 2021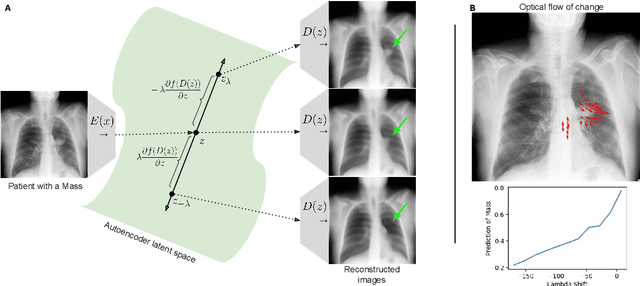

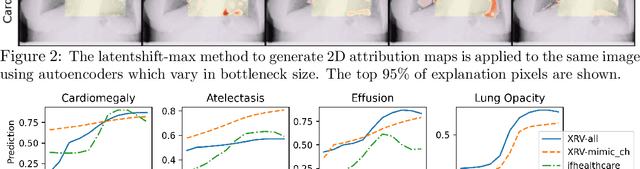
Abstract:Motivation: Traditional image attribution methods struggle to satisfactorily explain predictions of neural networks. Prediction explanation is important, especially in the medical imaging, for avoiding the unintended consequences of deploying AI systems when false positive predictions can impact patient care. Thus, there is a pressing need to develop improved models for model explainability and introspection. Specific Problem: A new approach is to transform input images to increase or decrease features which cause the prediction. However, current approaches are difficult to implement as they are monolithic or rely on GANs. These hurdles prevent wide adoption. Our approach: Given an arbitrary classifier, we propose a simple autoencoder and gradient update (Latent Shift) that can transform the latent representation of an input image to exaggerate or curtail the features used for prediction. We use this method to study chest X-ray classifiers and evaluate their performance. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to identify which ones are false positives (half are) using traditional attribution maps or our proposed method. Results: We found low overlap with ground truth pathology masks for models with reasonably high accuracy. However, the results from our reader study indicate that these models are generally looking at the correct features. We also found that the Latent Shift explanation allows a user to have more confidence in true positive predictions compared to traditional approaches (0.15$\pm$0.95 in a 5 point scale with p=0.01) with only a small increase in false positive predictions (0.04$\pm$1.06 with p=0.57). Accompanying webpage: https://mlmed.org/gifsplanation Source code: https://github.com/mlmed/gifsplanation
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge