Abdullah-Al-Zubaer Imran
Autoadaptive Medical Segment Anything Model
Jul 02, 2025Abstract:Medical image segmentation is a key task in the imaging workflow, influencing many image-based decisions. Traditional, fully-supervised segmentation models rely on large amounts of labeled training data, typically obtained through manual annotation, which can be an expensive, time-consuming, and error-prone process. This signals a need for accurate, automatic, and annotation-efficient methods of training these models. We propose ADA-SAM (automated, domain-specific, and adaptive segment anything model), a novel multitask learning framework for medical image segmentation that leverages class activation maps from an auxiliary classifier to guide the predictions of the semi-supervised segmentation branch, which is based on the Segment Anything (SAM) framework. Additionally, our ADA-SAM model employs a novel gradient feedback mechanism to create a learnable connection between the segmentation and classification branches by using the segmentation gradients to guide and improve the classification predictions. We validate ADA-SAM on real-world clinical data collected during rehabilitation trials, and demonstrate that our proposed method outperforms both fully-supervised and semi-supervised baselines by double digits in limited label settings. Our code is available at: https://github.com/tbwa233/ADA-SAM.
Domain and Task-Focused Example Selection for Data-Efficient Contrastive Medical Image Segmentation
May 25, 2025Abstract:Segmentation is one of the most important tasks in the medical imaging pipeline as it influences a number of image-based decisions. To be effective, fully supervised segmentation approaches require large amounts of manually annotated training data. However, the pixel-level annotation process is expensive, time-consuming, and error-prone, hindering progress and making it challenging to perform effective segmentations. Therefore, models must learn efficiently from limited labeled data. Self-supervised learning (SSL), particularly contrastive learning via pre-training on unlabeled data and fine-tuning on limited annotations, can facilitate such limited labeled image segmentation. To this end, we propose a novel self-supervised contrastive learning framework for medical image segmentation, leveraging inherent relationships of different images, dubbed PolyCL. Without requiring any pixel-level annotations or unreasonable data augmentations, our PolyCL learns and transfers context-aware discriminant features useful for segmentation from an innovative surrogate, in a task-related manner. Additionally, we integrate the Segment Anything Model (SAM) into our framework in two novel ways: as a post-processing refinement module that improves the accuracy of predicted masks using bounding box prompts derived from coarse outputs, and as a propagation mechanism via SAM 2 that generates volumetric segmentations from a single annotated 2D slice. Experimental evaluations on three public computed tomography (CT) datasets demonstrate that PolyCL outperforms fully-supervised and self-supervised baselines in both low-data and cross-domain scenarios. Our code is available at https://github.com/tbwa233/PolyCL.
Improving Brain Disorder Diagnosis with Advanced Brain Function Representation and Kolmogorov-Arnold Networks
Apr 04, 2025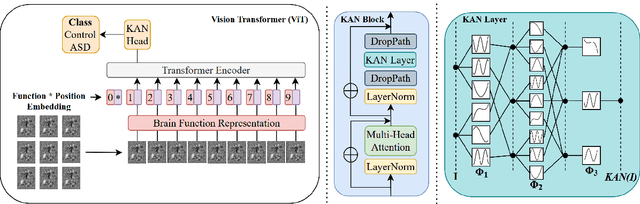
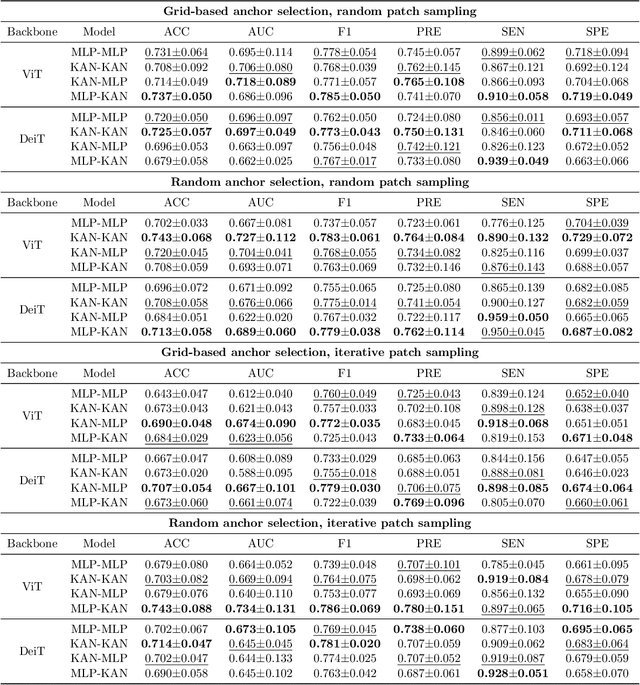
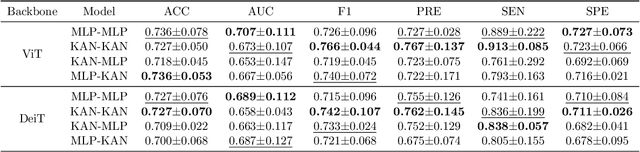
Abstract:Quantifying functional connectivity (FC), a vital metric for the diagnosis of various brain disorders, traditionally relies on the use of a pre-defined brain atlas. However, using such atlases can lead to issues regarding selection bias and lack of regard for specificity. Addressing this, we propose a novel transformer-based classification network (AFBR-KAN) with effective brain function representation to aid in diagnosing autism spectrum disorder (ASD). AFBR-KAN leverages Kolmogorov-Arnold Network (KAN) blocks replacing traditional multi-layer perceptron (MLP) components. Thorough experimentation reveals the effectiveness of AFBR-KAN in improving the diagnosis of ASD under various configurations of the model architecture. Our code is available at https://github.com/tbwa233/ABFR-KAN
Prompting Medical Vision-Language Models to Mitigate Diagnosis Bias by Generating Realistic Dermoscopic Images
Apr 02, 2025Abstract:Artificial Intelligence (AI) in skin disease diagnosis has improved significantly, but a major concern is that these models frequently show biased performance across subgroups, especially regarding sensitive attributes such as skin color. To address these issues, we propose a novel generative AI-based framework, namely, Dermatology Diffusion Transformer (DermDiT), which leverages text prompts generated via Vision Language Models and multimodal text-image learning to generate new dermoscopic images. We utilize large vision language models to generate accurate and proper prompts for each dermoscopic image which helps to generate synthetic images to improve the representation of underrepresented groups (patient, disease, etc.) in highly imbalanced datasets for clinical diagnoses. Our extensive experimentation showcases the large vision language models providing much more insightful representations, that enable DermDiT to generate high-quality images. Our code is available at https://github.com/Munia03/DermDiT
EarthScape: A Multimodal Dataset for Surficial Geologic Mapping and Earth Surface Analysis
Mar 19, 2025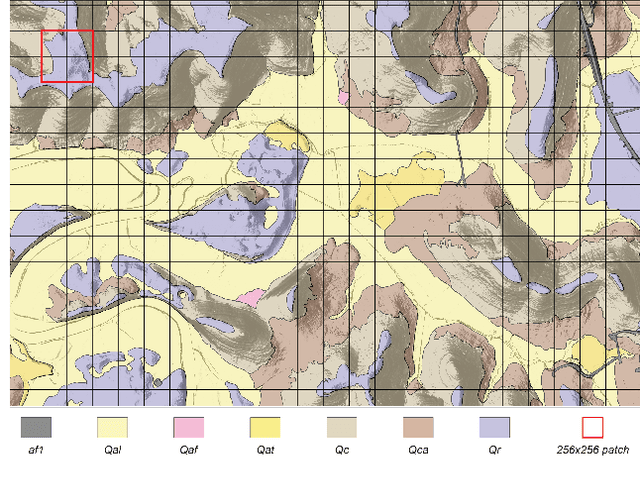
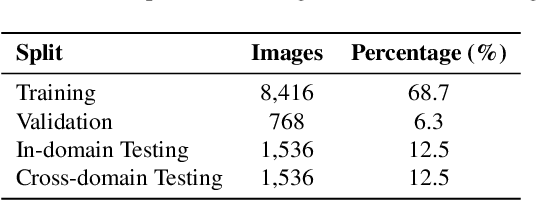
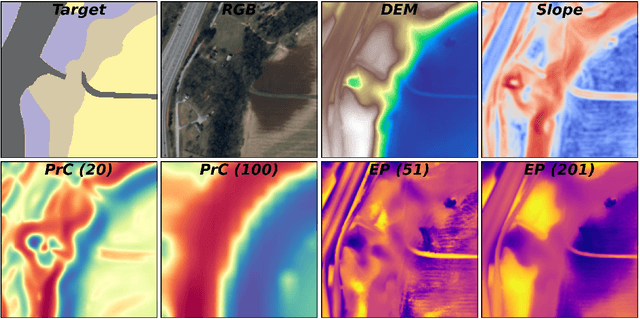
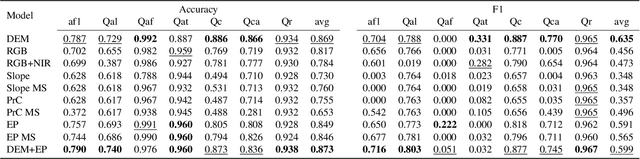
Abstract:Surficial geologic mapping is essential for understanding Earth surface processes, addressing modern challenges such as climate change and national security, and supporting common applications in engineering and resource management. However, traditional mapping methods are labor-intensive, limiting spatial coverage and introducing potential biases. To address these limitations, we introduce EarthScape, a novel, AI-ready multimodal dataset specifically designed for surficial geologic mapping and Earth surface analysis. EarthScape integrates high-resolution aerial RGB and near-infrared (NIR) imagery, digital elevation models (DEM), multi-scale DEM-derived terrain features, and hydrologic and infrastructure vector data. The dataset provides detailed annotations for seven distinct surficial geologic classes encompassing various geological processes. We present a comprehensive data processing pipeline using open-sourced raw data and establish baseline benchmarks using different spatial modalities to demonstrate the utility of EarthScape. As a living dataset with a vision for expansion, EarthScape bridges the gap between computer vision and Earth sciences, offering a valuable resource for advancing research in multimodal learning, geospatial analysis, and geological mapping. Our code is available at https://github.com/masseygeo/earthscape.
Annotation-Efficient Task Guidance for Medical Segment Anything
Dec 11, 2024



Abstract:Medical image segmentation is a key task in the imaging workflow, influencing many image-based decisions. Traditional, fully-supervised segmentation models rely on large amounts of labeled training data, typically obtained through manual annotation, which can be an expensive, time-consuming, and error-prone process. This signals a need for accurate, automatic, and annotation-efficient methods of training these models. We propose SAM-Mix, a novel multitask learning framework for medical image segmentation that uses class activation maps produced by an auxiliary classifier to guide the predictions of the semi-supervised segmentation branch, which is based on the SAM framework. Experimental evaluations on the public LiTS dataset confirm the effectiveness of SAM-Mix for simultaneous classification and segmentation of the liver from abdominal computed tomography (CT) scans. When trained for 90% fewer epochs on only 50 labeled 2D slices, representing just 0.04% of the available labeled training data, SAM-Mix achieves a Dice improvement of 5.1% over the best baseline model. The generalization results for SAM-Mix are even more impressive, with the same model configuration yielding a 25.4% Dice improvement on a cross-domain segmentation task. Our code is available at https://github.com/tbwa233/SAM-Mix.
Scout-Net: Prospective Personalized Estimation of CT Organ Doses from Scout Views
Dec 23, 2023Abstract:Purpose: Estimation of patient-specific organ doses is required for more comprehensive dose metrics, such as effective dose. Currently, available methods are performed retrospectively using the CT images themselves, which can only be done after the scan. To optimize CT acquisitions before scanning, rapid prediction of patient-specific organ dose is needed prospectively, using available scout images. We, therefore, devise an end-to-end, fully-automated deep learning solution to perform real-time, patient-specific, organ-level dosimetric estimation of CT scans. Approach: We propose the Scout-Net model for CT dose prediction at six different organs as well as for the overall patient body, leveraging the routinely obtained frontal and lateral scout images of patients, before their CT scans. To obtain reference values of the organ doses, we used Monte Carlo simulation and 3D segmentation methods on the corresponding CT images of the patients. Results: We validate our proposed Scout-Net model against real patient CT data and demonstrate the effectiveness in estimating organ doses in real-time (only 27 ms on average per scan). Additionally, we demonstrate the efficiency (real-time execution), sufficiency (reasonable error rates), and robustness (consistent across varying patient sizes) of the Scout-Net model. Conclusions: An effective, efficient, and robust Scout-Net model, once incorporated into the CT acquisition plan, could potentially guide the automatic exposure control for balanced image quality and radiation dose.
Forward-Forward Contrastive Learning
May 04, 2023Abstract:Medical image classification is one of the most important tasks for computer-aided diagnosis. Deep learning models, particularly convolutional neural networks, have been successfully used for disease classification from medical images, facilitated by automated feature learning. However, the diverse imaging modalities and clinical pathology make it challenging to construct generalized and robust classifications. Towards improving the model performance, we propose a novel pretraining approach, namely Forward Forward Contrastive Learning (FFCL), which leverages the Forward-Forward Algorithm in a contrastive learning framework--both locally and globally. Our experimental results on the chest X-ray dataset indicate that the proposed FFCL achieves superior performance (3.69% accuracy over ImageNet pretrained ResNet-18) over existing pretraining models in the pneumonia classification task. Moreover, extensive ablation experiments support the particular local and global contrastive pretraining design in FFCL.
Semi-Supervised Relational Contrastive Learning
Apr 11, 2023Abstract:Disease diagnosis from medical images via supervised learning is usually dependent on tedious, error-prone, and costly image labeling by medical experts. Alternatively, semi-supervised learning and self-supervised learning offer effectiveness through the acquisition of valuable insights from readily available unlabeled images. We present Semi-Supervised Relational Contrastive Learning (SRCL), a novel semi-supervised learning model that leverages self-supervised contrastive loss and sample relation consistency for the more meaningful and effective exploitation of unlabeled data. Our experimentation with the SRCL model explores both pre-train/fine-tune and joint learning of the pretext (contrastive learning) and downstream (diagnostic classification) tasks. We validate against the ISIC 2018 Challenge benchmark skin lesion classification dataset and demonstrate the effectiveness of our semi-supervised method on varying amounts of labeled data.
Single Image Internal Distribution Measurement Using Non-Local Variational Autoencoder
Apr 02, 2022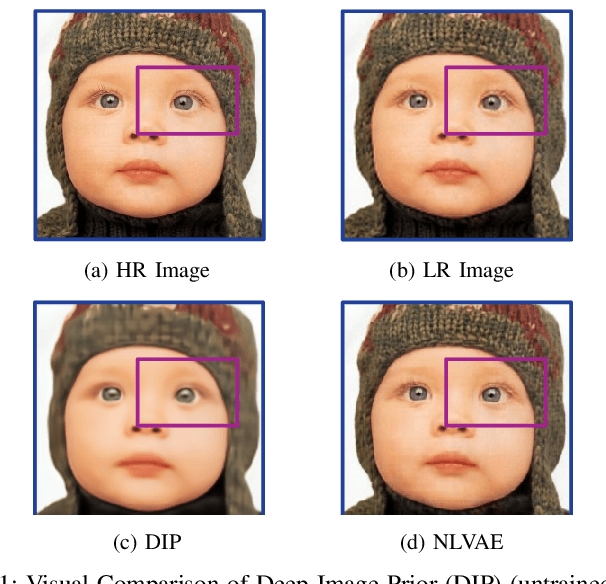
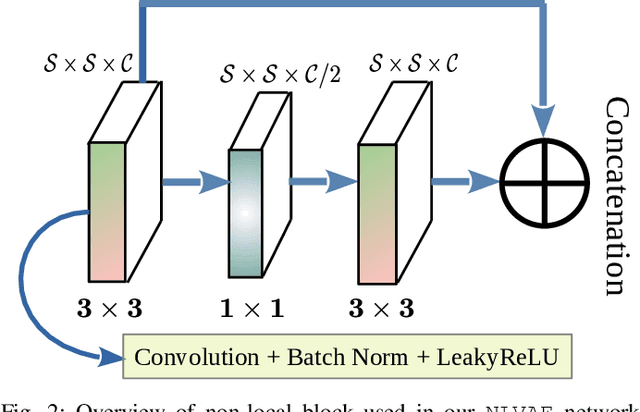
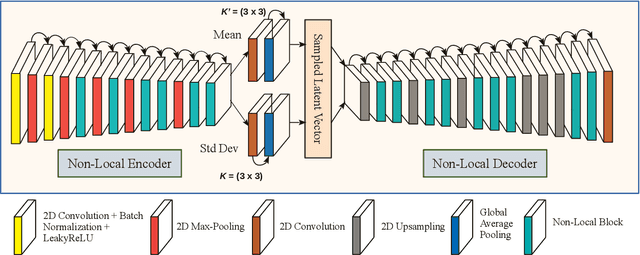
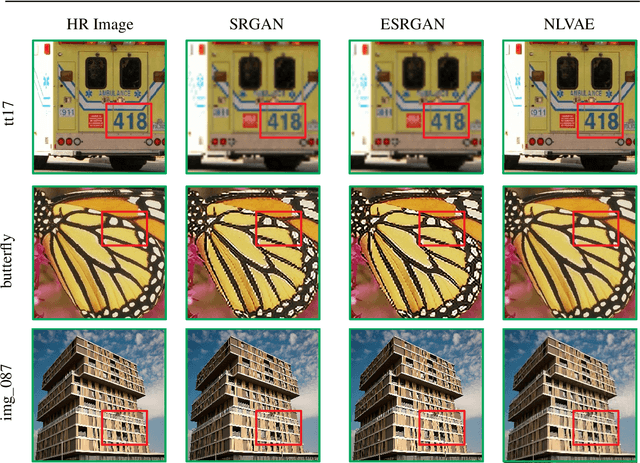
Abstract:Deep learning-based super-resolution methods have shown great promise, especially for single image super-resolution (SISR) tasks. Despite the performance gain, these methods are limited due to their reliance on copious data for model training. In addition, supervised SISR solutions rely on local neighbourhood information focusing only on the feature learning processes for the reconstruction of low-dimensional images. Moreover, they fail to capitalize on global context due to their constrained receptive field. To combat these challenges, this paper proposes a novel image-specific solution, namely non-local variational autoencoder (\texttt{NLVAE}), to reconstruct a high-resolution (HR) image from a single low-resolution (LR) image without the need for any prior training. To harvest maximum details for various receptive regions and high-quality synthetic images, \texttt{NLVAE} is introduced as a self-supervised strategy that reconstructs high-resolution images using disentangled information from the non-local neighbourhood. Experimental results from seven benchmark datasets demonstrate the effectiveness of the \texttt{NLVAE} model. Moreover, our proposed model outperforms a number of baseline and state-of-the-art methods as confirmed through extensive qualitative and quantitative evaluations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge