Adam Wang
Poisson flow consistency models for low-dose CT image denoising
Feb 13, 2024



Abstract:Diffusion and Poisson flow models have demonstrated remarkable success for a wide range of generative tasks. Nevertheless, their iterative nature results in computationally expensive sampling and the number of function evaluations (NFE) required can be orders of magnitude larger than for single-step methods. Consistency models are a recent class of deep generative models which enable single-step sampling of high quality data without the need for adversarial training. In this paper, we introduce a novel image denoising technique which combines the flexibility afforded in Poisson flow generative models (PFGM)++ with the, high quality, single step sampling of consistency models. The proposed method first learns a trajectory between a noise distribution and the posterior distribution of interest by training PFGM++ in a supervised fashion. These pre-trained PFGM++ are subsequently "distilled" into Poisson flow consistency models (PFCM) via an updated version of consistency distillation. We call this approach posterior sampling Poisson flow consistency models (PS-PFCM). Our results indicate that the added flexibility of tuning the hyperparameter D, the dimensionality of the augmentation variables in PFGM++, allows us to outperform consistency models, a current state-of-the-art diffusion-style model with NFE=1 on clinical low-dose CT images. Notably, PFCM is in itself a novel family of deep generative models and we provide initial results on the CIFAR-10 dataset.
Scout-Net: Prospective Personalized Estimation of CT Organ Doses from Scout Views
Dec 23, 2023Abstract:Purpose: Estimation of patient-specific organ doses is required for more comprehensive dose metrics, such as effective dose. Currently, available methods are performed retrospectively using the CT images themselves, which can only be done after the scan. To optimize CT acquisitions before scanning, rapid prediction of patient-specific organ dose is needed prospectively, using available scout images. We, therefore, devise an end-to-end, fully-automated deep learning solution to perform real-time, patient-specific, organ-level dosimetric estimation of CT scans. Approach: We propose the Scout-Net model for CT dose prediction at six different organs as well as for the overall patient body, leveraging the routinely obtained frontal and lateral scout images of patients, before their CT scans. To obtain reference values of the organ doses, we used Monte Carlo simulation and 3D segmentation methods on the corresponding CT images of the patients. Results: We validate our proposed Scout-Net model against real patient CT data and demonstrate the effectiveness in estimating organ doses in real-time (only 27 ms on average per scan). Additionally, we demonstrate the efficiency (real-time execution), sufficiency (reasonable error rates), and robustness (consistent across varying patient sizes) of the Scout-Net model. Conclusions: An effective, efficient, and robust Scout-Net model, once incorporated into the CT acquisition plan, could potentially guide the automatic exposure control for balanced image quality and radiation dose.
Coronary Atherosclerotic Plaque Characterization with Photon-counting CT: a Simulation-based Feasibility Study
Dec 04, 2023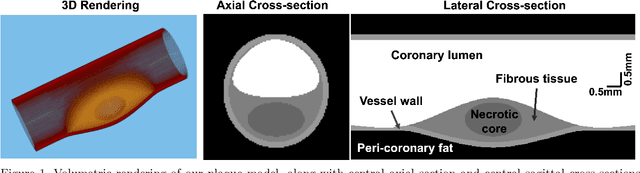
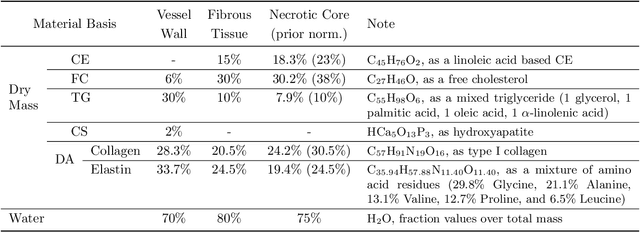
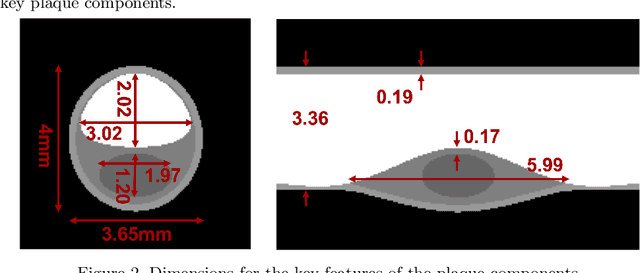
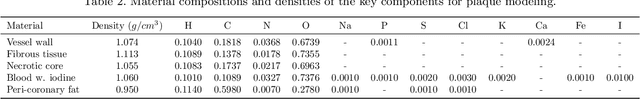
Abstract:Recent development of photon-counting CT (PCCT) brings great opportunities for plaque characterization with much-improved spatial resolution and spectral imaging capability. While existing coronary plaque PCCT imaging results are based on detectors made of CZT or CdTe materials, deep-silicon photon-counting detectors have unique performance characteristics and promise distinct imaging capabilities. In this work, we report a systematic simulation study of a deep-silicon PCCT scanner with a new clinically-relevant digital plaque phantom with realistic geometrical parameters and chemical compositions. This work investigates the effects of spatial resolution, noise, motion artifacts, radiation dose, and spectral characterization. Our simulation results suggest that the deep-silicon PCCT design provides adequate spatial resolution for visualizing a necrotic core and quantitation of key plaque features. Advanced denoising techniques and aggressive bowtie filter designs can keep image noise to acceptable levels at this resolution while keeping radiation dose comparable to that of a conventional CT scan. The ultrahigh resolution of PCCT also means an elevated sensitivity to motion artifacts. It is found that a tolerance of less than 0.4 mm residual movement range requires the application of accurate motion correction methods for best plaque imaging quality with PCCT.
Fairness-enhancing mixed effects deep learning improves fairness on in- and out-of-distribution clustered (non-iid) data
Oct 04, 2023Abstract:Traditional deep learning (DL) suffers from two core problems. Firstly, it assumes training samples are independent and identically distributed. However, numerous real-world datasets group samples by shared measurements (e.g., study participants or cells), violating this assumption. In these scenarios, DL can show compromised performance, limited generalization, and interpretability issues, coupled with cluster confounding causing Type 1 and 2 errors. Secondly, models are typically trained for overall accuracy, often neglecting underrepresented groups and introducing biases in crucial areas like loan approvals or determining health insurance rates, such biases can significantly impact one's quality of life. To address both of these challenges simultaneously, we present a mixed effects deep learning (MEDL) framework. MEDL separately quantifies cluster-invariant fixed effects (FE) and cluster-specific random effects (RE) through the introduction of: 1) a cluster adversary which encourages the learning of cluster-invariant FE, 2) a Bayesian neural network which quantifies the RE, and a mixing function combining the FE an RE into a mixed-effect prediction. We marry this MEDL with adversarial debiasing, which promotes equality-of-odds fairness across FE, RE, and ME predictions for fairness-sensitive variables. We evaluated our approach using three datasets: two from census/finance focusing on income classification and one from healthcare predicting hospitalization duration, a regression task. Our framework notably enhances fairness across all sensitive variables-increasing fairness up to 82% for age, 43% for race, 86% for sex, and 27% for marital-status. Besides promoting fairness, our method maintains the robust performance and clarity of MEDL. It's versatile, suitable for various dataset types and tasks, making it broadly applicable. Our GitHub repository houses the implementation.
Semi-Supervised Relational Contrastive Learning
Apr 11, 2023Abstract:Disease diagnosis from medical images via supervised learning is usually dependent on tedious, error-prone, and costly image labeling by medical experts. Alternatively, semi-supervised learning and self-supervised learning offer effectiveness through the acquisition of valuable insights from readily available unlabeled images. We present Semi-Supervised Relational Contrastive Learning (SRCL), a novel semi-supervised learning model that leverages self-supervised contrastive loss and sample relation consistency for the more meaningful and effective exploitation of unlabeled data. Our experimentation with the SRCL model explores both pre-train/fine-tune and joint learning of the pretext (contrastive learning) and downstream (diagnostic classification) tasks. We validate against the ISIC 2018 Challenge benchmark skin lesion classification dataset and demonstrate the effectiveness of our semi-supervised method on varying amounts of labeled data.
Generalized Multi-Task Learning from Substantially Unlabeled Multi-Source Medical Image Data
Oct 25, 2021



Abstract:Deep learning-based models, when trained in a fully-supervised manner, can be effective in performing complex image analysis tasks, although contingent upon the availability of large labeled datasets. Especially in the medical imaging domain, however, expert image annotation is expensive, time-consuming, and prone to variability. Semi-supervised learning from limited quantities of labeled data has shown promise as an alternative. Maximizing knowledge gains from copious unlabeled data benefits semi-supervised learning models. Moreover, learning multiple tasks within the same model further improves its generalizability. We propose MultiMix, a new multi-task learning model that jointly learns disease classification and anatomical segmentation in a semi-supervised manner, while preserving explainability through a novel saliency bridge between the two tasks. Our experiments with varying quantities of multi-source labeled data in the training sets confirm the effectiveness of MultiMix in the simultaneous classification of pneumonia and segmentation of the lungs in chest X-ray images. Moreover, both in-domain and cross-domain evaluations across these tasks further showcase the potential of our model to adapt to challenging generalization scenarios.
Window-Level is a Strong Denoising Surrogate
May 15, 2021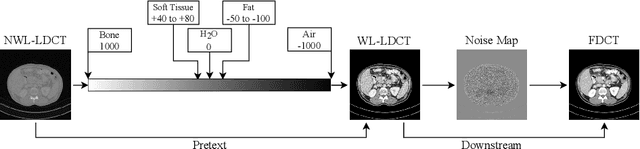

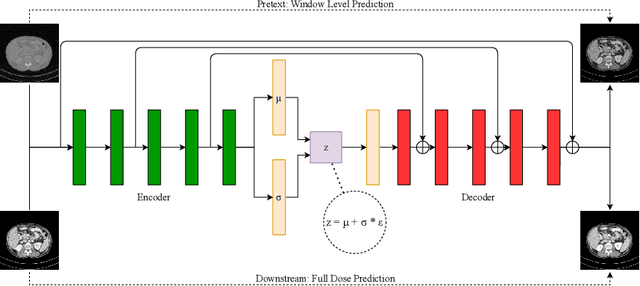
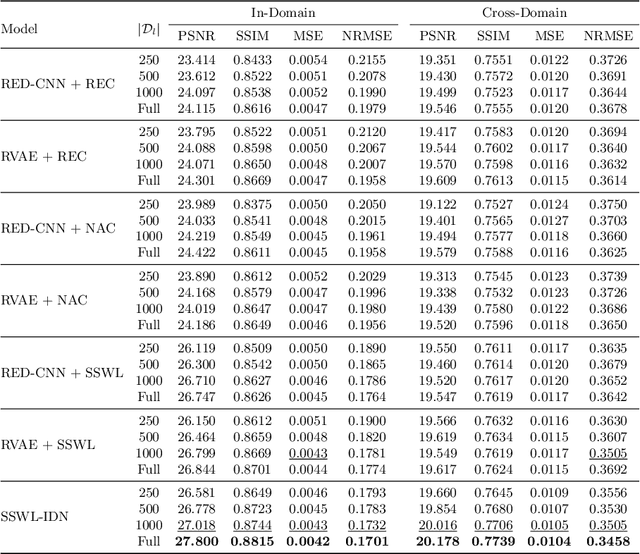
Abstract:CT image quality is heavily reliant on radiation dose, which causes a trade-off between radiation dose and image quality that affects the subsequent image-based diagnostic performance. However, high radiation can be harmful to both patients and operators. Several (deep learning-based) approaches have been attempted to denoise low dose images. However, those approaches require access to large training sets, specifically the full dose CT images for reference, which can often be difficult to obtain. Self-supervised learning is an emerging alternative for lowering the reference data requirement facilitating unsupervised learning. Currently available self-supervised CT denoising works are either dependent on foreign domain or pretexts are not very task-relevant. To tackle the aforementioned challenges, we propose a novel self-supervised learning approach, namely Self-Supervised Window-Leveling for Image DeNoising (SSWL-IDN), leveraging an innovative, task-relevant, simple, yet effective surrogate -- prediction of the window-leveled equivalent. SSWL-IDN leverages residual learning and a hybrid loss combining perceptual loss and MSE, all incorporated in a VAE framework. Our extensive (in- and cross-domain) experimentation demonstrates the effectiveness of SSWL-IDN in aggressive denoising of CT (abdomen and chest) images acquired at 5\% dose level only.
MultiMix: Sparingly Supervised, Extreme Multitask Learning From Medical Images
Oct 28, 2020



Abstract:Semi-supervised learning via learning from limited quantities of labeled data has been investigated as an alternative to supervised counterparts. Maximizing knowledge gains from copious unlabeled data benefit semi-supervised learning settings. Moreover, learning multiple tasks within the same model further improves model generalizability. We propose a novel multitask learning model, namely MultiMix, which jointly learns disease classification and anatomical segmentation in a sparingly supervised manner, while preserving explainability through bridge saliency between the two tasks. Our extensive experimentation with varied quantities of labeled data in the training sets justify the effectiveness of our multitasking model for the classification of pneumonia and segmentation of lungs from chest X-ray images. Moreover, both in-domain and cross-domain evaluations across the tasks further showcase the potential of our model to adapt to challenging generalization scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge