Anuj Pareek
Evaluating and Improving the Effectiveness of Synthetic Chest X-Rays for Medical Image Analysis
Nov 27, 2024Abstract:Purpose: To explore best-practice approaches for generating synthetic chest X-ray images and augmenting medical imaging datasets to optimize the performance of deep learning models in downstream tasks like classification and segmentation. Materials and Methods: We utilized a latent diffusion model to condition the generation of synthetic chest X-rays on text prompts and/or segmentation masks. We explored methods like using a proxy model and using radiologist feedback to improve the quality of synthetic data. These synthetic images were then generated from relevant disease information or geometrically transformed segmentation masks and added to ground truth training set images from the CheXpert, CANDID-PTX, SIIM, and RSNA Pneumonia datasets to measure improvements in classification and segmentation model performance on the test sets. F1 and Dice scores were used to evaluate classification and segmentation respectively. One-tailed t-tests with Bonferroni correction assessed the statistical significance of performance improvements with synthetic data. Results: Across all experiments, the synthetic data we generated resulted in a maximum mean classification F1 score improvement of 0.150453 (CI: 0.099108-0.201798; P=0.0031) compared to using only real data. For segmentation, the maximum Dice score improvement was 0.14575 (CI: 0.108267-0.183233; P=0.0064). Conclusion: Best practices for generating synthetic chest X-ray images for downstream tasks include conditioning on single-disease labels or geometrically transformed segmentation masks, as well as potentially using proxy modeling for fine-tuning such generations.
Clinical Text Summarization: Adapting Large Language Models Can Outperform Human Experts
Sep 14, 2023Abstract:Sifting through vast textual data and summarizing key information imposes a substantial burden on how clinicians allocate their time. Although large language models (LLMs) have shown immense promise in natural language processing (NLP) tasks, their efficacy across diverse clinical summarization tasks has not yet been rigorously examined. In this work, we employ domain adaptation methods on eight LLMs, spanning six datasets and four distinct summarization tasks: radiology reports, patient questions, progress notes, and doctor-patient dialogue. Our thorough quantitative assessment reveals trade-offs between models and adaptation methods in addition to instances where recent advances in LLMs may not lead to improved results. Further, in a clinical reader study with six physicians, we depict that summaries from the best adapted LLM are preferable to human summaries in terms of completeness and correctness. Our ensuing qualitative analysis delineates mutual challenges faced by both LLMs and human experts. Lastly, we correlate traditional quantitative NLP metrics with reader study scores to enhance our understanding of how these metrics align with physician preferences. Our research marks the first evidence of LLMs outperforming human experts in clinical text summarization across multiple tasks. This implies that integrating LLMs into clinical workflows could alleviate documentation burden, empowering clinicians to focus more on personalized patient care and other irreplaceable human aspects of medicine.
RadAdapt: Radiology Report Summarization via Lightweight Domain Adaptation of Large Language Models
May 02, 2023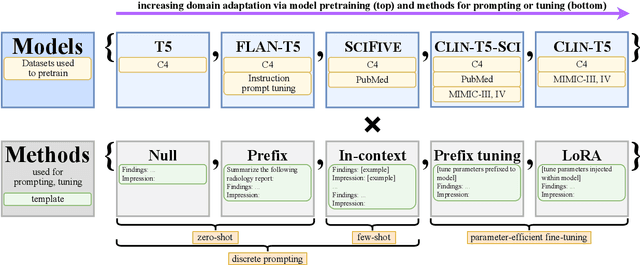
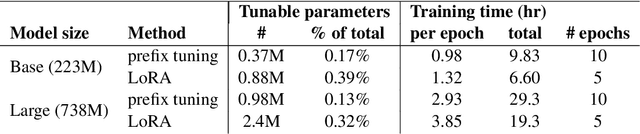
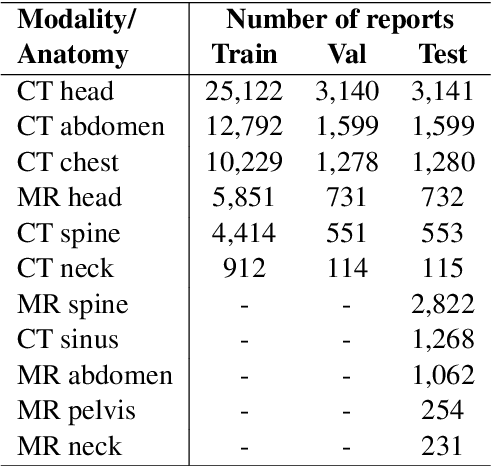

Abstract:We systematically investigate lightweight strategies to adapt large language models (LLMs) for the task of radiology report summarization (RRS). Specifically, we focus on domain adaptation via pretraining (on natural language, biomedical text, and clinical text) and via prompting (zero-shot, in-context learning) or parameter-efficient fine-tuning (prefix tuning, LoRA). Our results on the MIMIC-III dataset consistently demonstrate best performance by maximally adapting to the task via pretraining on clinical text and parameter-efficient fine-tuning on RRS examples. Importantly, this method fine-tunes a mere 0.32% of parameters throughout the model, in contrast to end-to-end fine-tuning (100% of parameters). Additionally, we study the effect of in-context examples and out-of-distribution (OOD) training before concluding with a radiologist reader study and qualitative analysis. Our findings highlight the importance of domain adaptation in RRS and provide valuable insights toward developing effective natural language processing solutions for clinical tasks.
The Effect of Counterfactuals on Reading Chest X-rays
Apr 02, 2023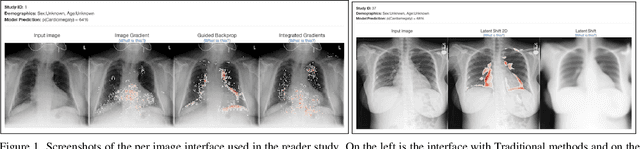
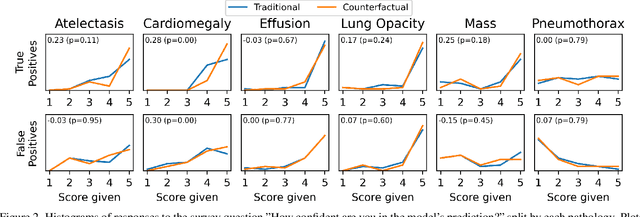
Abstract:This study evaluates the effect of counterfactual explanations on the interpretation of chest X-rays. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to rate their confidence that the model's prediction is correct using a 5 point scale. Half of the predictions are false positives. Each prediction is explained twice, once using traditional attribution methods and once with a counterfactual explanation. The overall results indicate that counterfactual explanations allow a radiologist to have more confidence in true positive predictions compared to traditional approaches (0.15$\pm$0.95 with p=0.01) with only a small increase in false positive predictions (0.04$\pm$1.06 with p=0.57). We observe the specific prediction tasks of Mass and Atelectasis appear to benefit the most compared to other tasks.
CheXternal: Generalization of Deep Learning Models for Chest X-ray Interpretation to Photos of Chest X-rays and External Clinical Settings
Feb 21, 2021



Abstract:Recent advances in training deep learning models have demonstrated the potential to provide accurate chest X-ray interpretation and increase access to radiology expertise. However, poor generalization due to data distribution shifts in clinical settings is a key barrier to implementation. In this study, we measured the diagnostic performance for 8 different chest X-ray models when applied to (1) smartphone photos of chest X-rays and (2) external datasets without any finetuning. All models were developed by different groups and submitted to the CheXpert challenge, and re-applied to test datasets without further tuning. We found that (1) on photos of chest X-rays, all 8 models experienced a statistically significant drop in task performance, but only 3 performed significantly worse than radiologists on average, and (2) on the external set, none of the models performed statistically significantly worse than radiologists, and five models performed statistically significantly better than radiologists. Our results demonstrate that some chest X-ray models, under clinically relevant distribution shifts, were comparable to radiologists while other models were not. Future work should investigate aspects of model training procedures and dataset collection that influence generalization in the presence of data distribution shifts.
Gifsplanation via Latent Shift: A Simple Autoencoder Approach to Progressive Exaggeration on Chest X-rays
Feb 18, 2021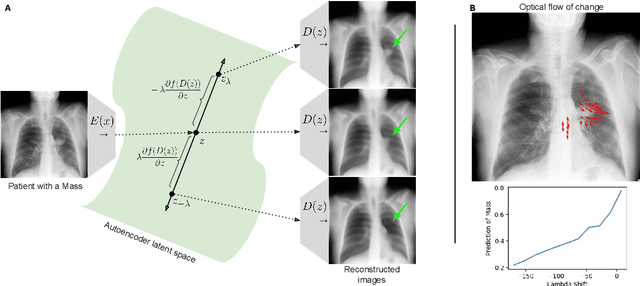

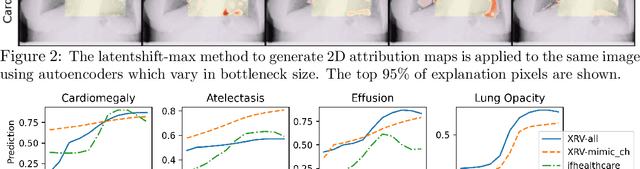
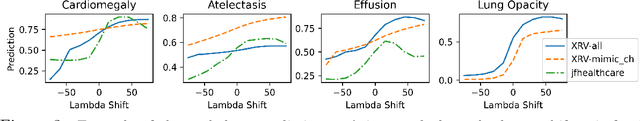
Abstract:Motivation: Traditional image attribution methods struggle to satisfactorily explain predictions of neural networks. Prediction explanation is important, especially in the medical imaging, for avoiding the unintended consequences of deploying AI systems when false positive predictions can impact patient care. Thus, there is a pressing need to develop improved models for model explainability and introspection. Specific Problem: A new approach is to transform input images to increase or decrease features which cause the prediction. However, current approaches are difficult to implement as they are monolithic or rely on GANs. These hurdles prevent wide adoption. Our approach: Given an arbitrary classifier, we propose a simple autoencoder and gradient update (Latent Shift) that can transform the latent representation of an input image to exaggerate or curtail the features used for prediction. We use this method to study chest X-ray classifiers and evaluate their performance. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to identify which ones are false positives (half are) using traditional attribution maps or our proposed method. Results: We found low overlap with ground truth pathology masks for models with reasonably high accuracy. However, the results from our reader study indicate that these models are generally looking at the correct features. We also found that the Latent Shift explanation allows a user to have more confidence in true positive predictions compared to traditional approaches (0.15$\pm$0.95 in a 5 point scale with p=0.01) with only a small increase in false positive predictions (0.04$\pm$1.06 with p=0.57). Accompanying webpage: https://mlmed.org/gifsplanation Source code: https://github.com/mlmed/gifsplanation
CheXphotogenic: Generalization of Deep Learning Models for Chest X-ray Interpretation to Photos of Chest X-rays
Nov 12, 2020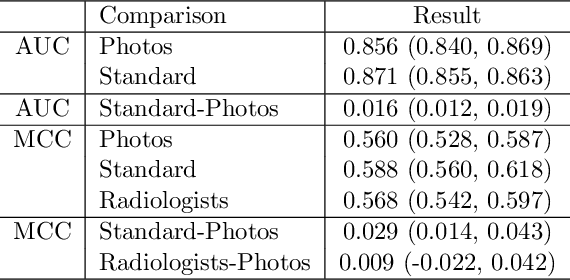
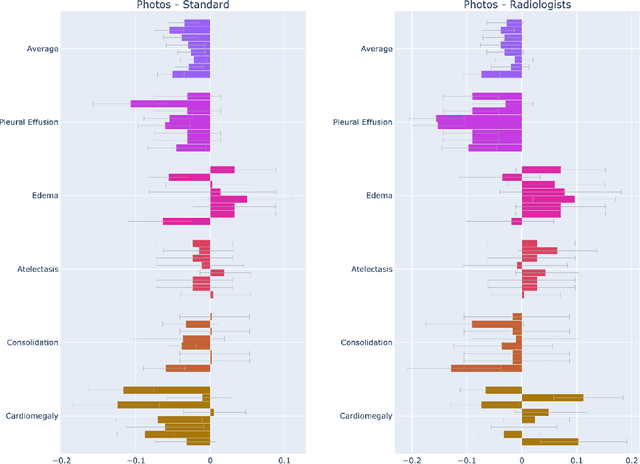
Abstract:The use of smartphones to take photographs of chest x-rays represents an appealing solution for scaled deployment of deep learning models for chest x-ray interpretation. However, the performance of chest x-ray algorithms on photos of chest x-rays has not been thoroughly investigated. In this study, we measured the diagnostic performance for 8 different chest x-ray models when applied to photos of chest x-rays. All models were developed by different groups and submitted to the CheXpert challenge, and re-applied to smartphone photos of x-rays in the CheXphoto dataset without further tuning. We found that several models had a drop in performance when applied to photos of chest x-rays, but even with this drop, some models still performed comparably to radiologists. Further investigation could be directed towards understanding how different model training procedures may affect model generalization to photos of chest x-rays.
CheXphoto: 10,000+ Smartphone Photos and Synthetic Photographic Transformations of Chest X-rays for Benchmarking Deep Learning Robustness
Jul 13, 2020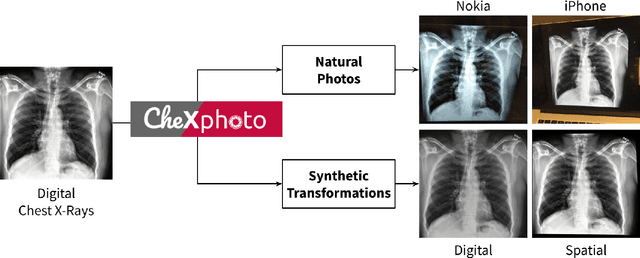
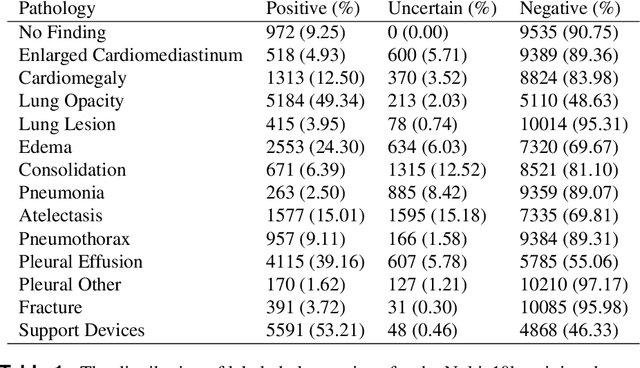
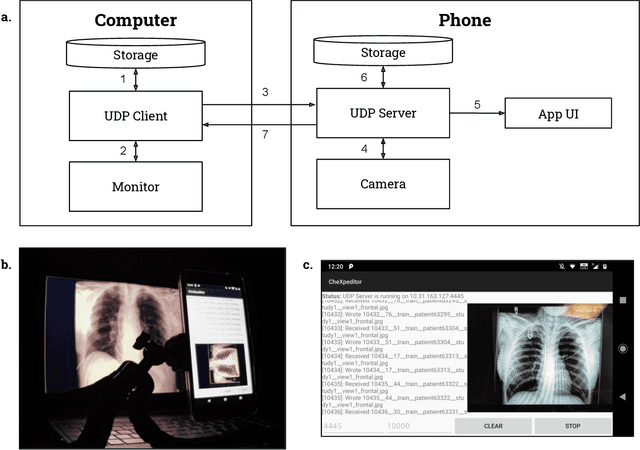
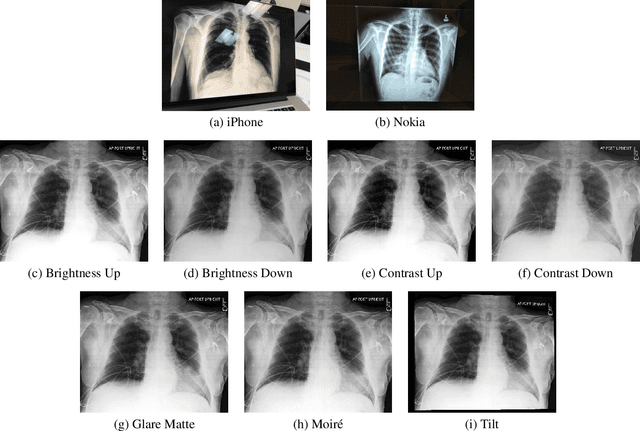
Abstract:Clinical deployment of deep learning algorithms for chest x-ray interpretation requires a solution that can integrate into the vast spectrum of clinical workflows across the world. An appealing solution to scaled deployment is to leverage the existing ubiquity of smartphones: in several parts of the world, clinicians and radiologists capture photos of chest x-rays to share with other experts or clinicians via smartphone using messaging services like WhatsApp. However, the application of chest x-ray algorithms to photos of chest x-rays requires reliable classification in the presence of smartphone photo artifacts such as screen glare and poor viewing angle not typically encountered on digital x-rays used to train machine learning models. We introduce CheXphoto, a dataset of smartphone photos and synthetic photographic transformations of chest x-rays sampled from the CheXpert dataset. To generate CheXphoto we (1) automatically and manually captured photos of digital x-rays under different settings, including various lighting conditions and locations, and, (2) generated synthetic transformations of digital x-rays targeted to make them look like photos of digital x-rays and x-ray films. We release this dataset as a resource for testing and improving the robustness of deep learning algorithms for automated chest x-ray interpretation on smartphone photos of chest x-rays.
CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT
Apr 30, 2020



Abstract:The extraction of labels from radiology text reports enables large-scale training of medical imaging models. Existing approaches to report labeling typically rely either on sophisticated feature engineering based on medical domain knowledge or manual annotations by experts. In this work, we introduce a BERT-based approach to medical image report labeling that exploits both the scale of available rule-based systems and the quality of expert annotations. We demonstrate superior performance of a biomedically pretrained BERT model first trained on annotations of a rule-based labeler and then finetuned on a small set of expert annotations augmented with automated backtranslation. We find that our final model, CheXbert, is able to outperform the previous best rules-based labeler with statistical significance, setting a new SOTA for report labeling on one of the largest datasets of chest x-rays.
CheXpedition: Investigating Generalization Challenges for Translation of Chest X-Ray Algorithms to the Clinical Setting
Mar 11, 2020
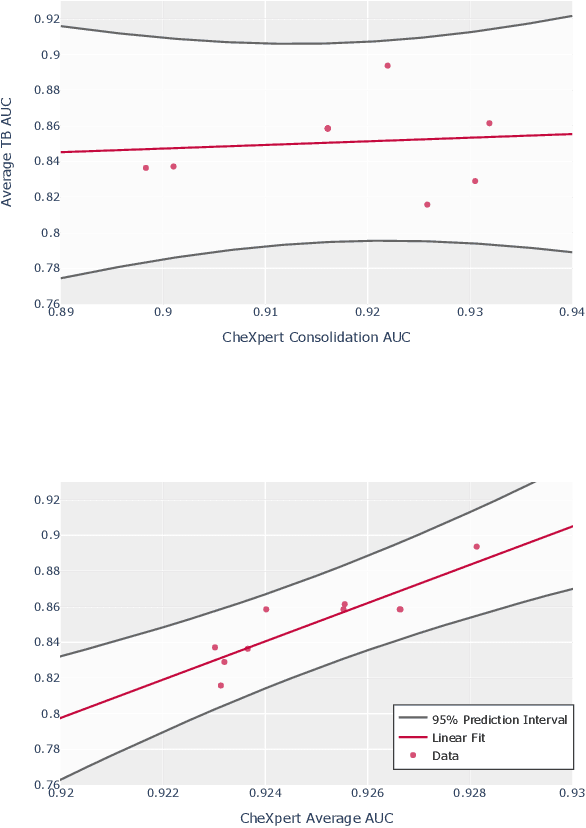
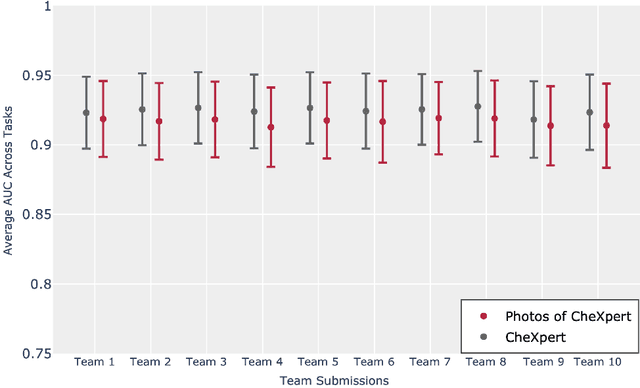
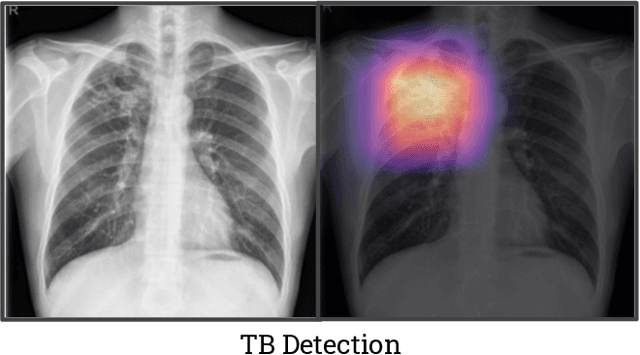
Abstract:Although there have been several recent advances in the application of deep learning algorithms to chest x-ray interpretation, we identify three major challenges for the translation of chest x-ray algorithms to the clinical setting. We examine the performance of the top 10 performing models on the CheXpert challenge leaderboard on three tasks: (1) TB detection, (2) pathology detection on photos of chest x-rays, and (3) pathology detection on data from an external institution. First, we find that the top 10 chest x-ray models on the CheXpert competition achieve an average AUC of 0.851 on the task of detecting TB on two public TB datasets without fine-tuning or including the TB labels in training data. Second, we find that the average performance of the models on photos of x-rays (AUC = 0.916) is similar to their performance on the original chest x-ray images (AUC = 0.924). Third, we find that the models tested on an external dataset either perform comparably to or exceed the average performance of radiologists. We believe that our investigation will inform rapid translation of deep learning algorithms to safe and effective clinical decision support tools that can be validated prospectively with large impact studies and clinical trials.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge