Phil Chen
Evidential Softmax for Sparse Multimodal Distributions in Deep Generative Models
Oct 27, 2021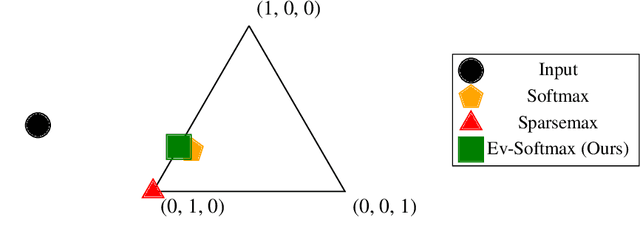
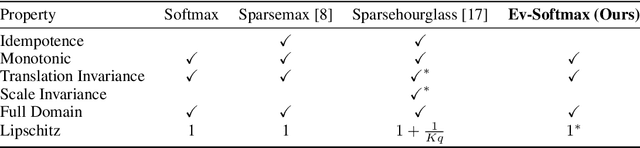
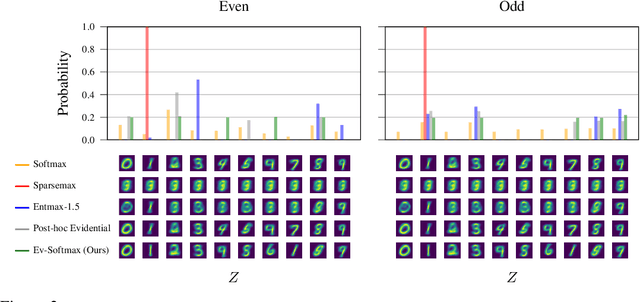
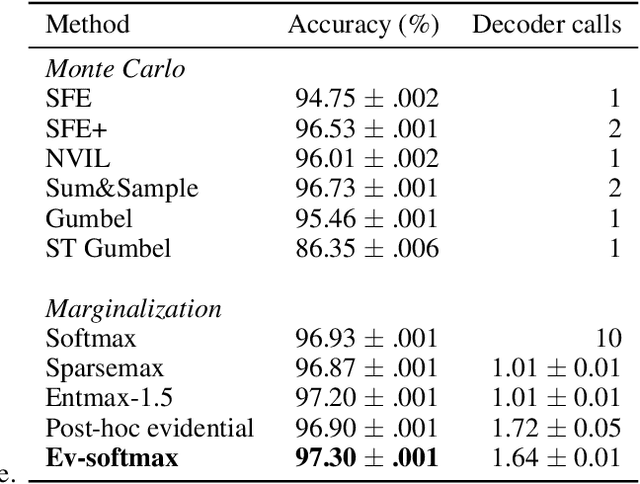
Abstract:Many applications of generative models rely on the marginalization of their high-dimensional output probability distributions. Normalization functions that yield sparse probability distributions can make exact marginalization more computationally tractable. However, sparse normalization functions usually require alternative loss functions for training since the log-likelihood is undefined for sparse probability distributions. Furthermore, many sparse normalization functions often collapse the multimodality of distributions. In this work, we present $\textit{ev-softmax}$, a sparse normalization function that preserves the multimodality of probability distributions. We derive its properties, including its gradient in closed-form, and introduce a continuous family of approximations to $\textit{ev-softmax}$ that have full support and can be trained with probabilistic loss functions such as negative log-likelihood and Kullback-Leibler divergence. We evaluate our method on a variety of generative models, including variational autoencoders and auto-regressive architectures. Our method outperforms existing dense and sparse normalization techniques in distributional accuracy. We demonstrate that $\textit{ev-softmax}$ successfully reduces the dimensionality of probability distributions while maintaining multimodality.
CheXpedition: Investigating Generalization Challenges for Translation of Chest X-Ray Algorithms to the Clinical Setting
Mar 11, 2020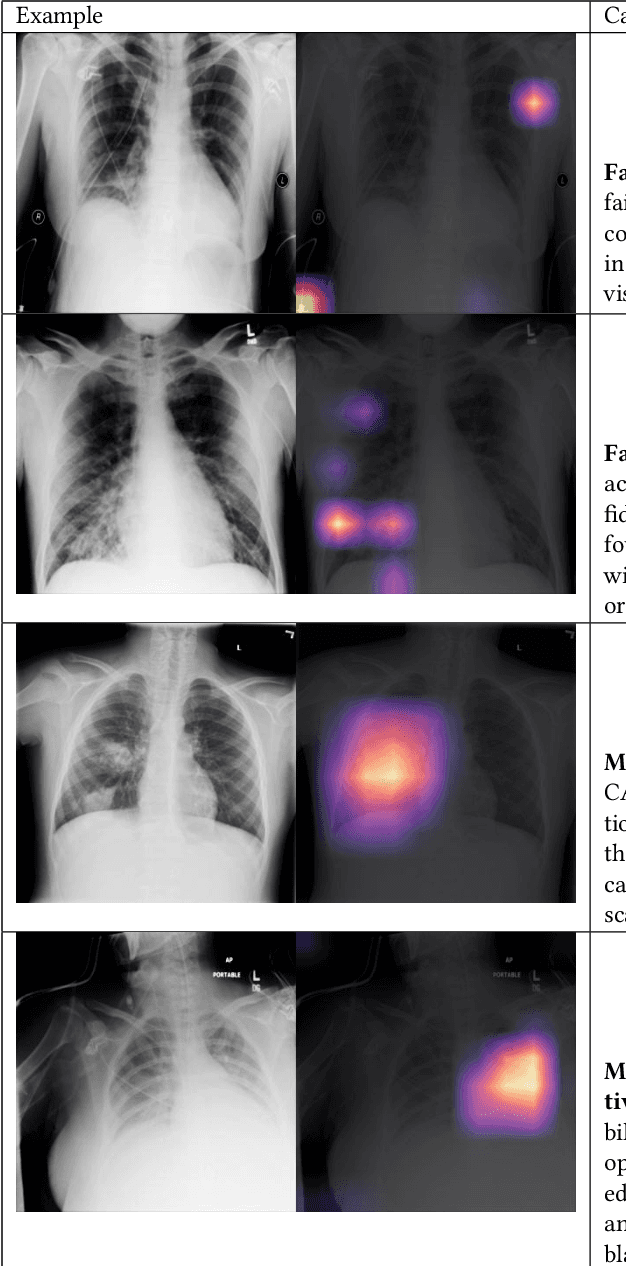
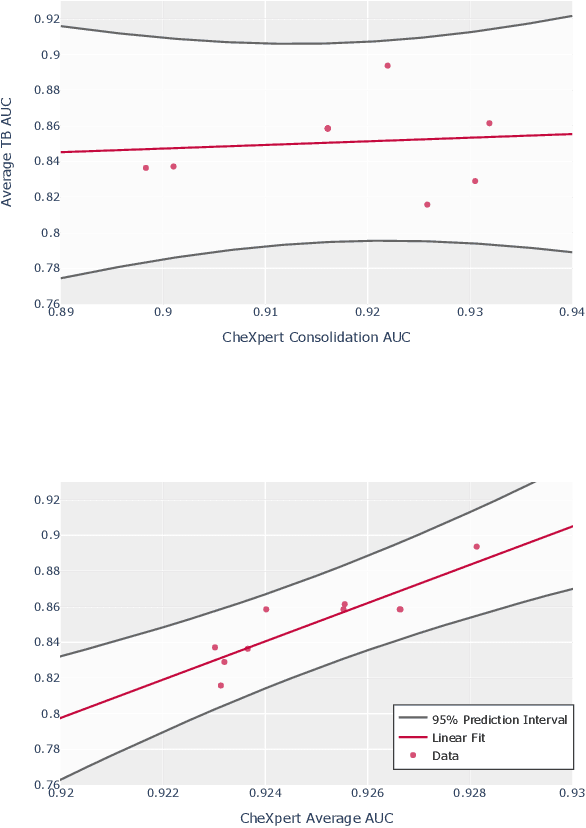
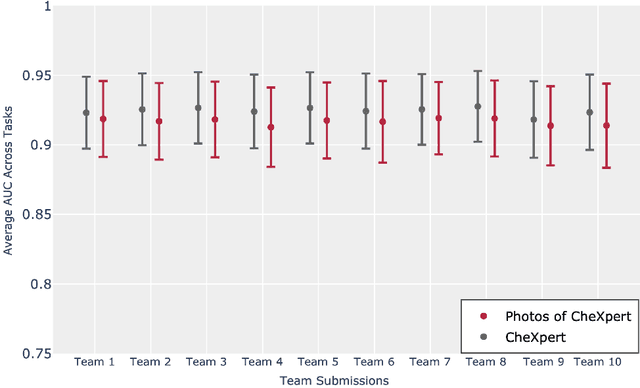
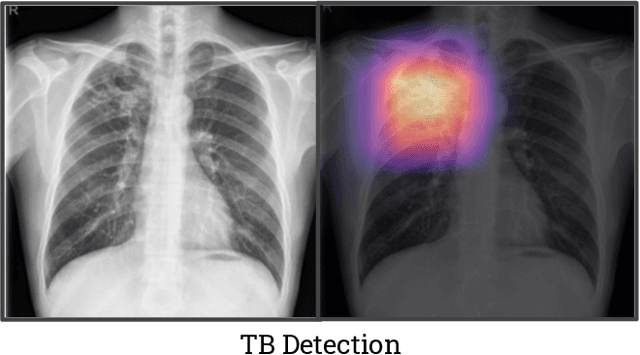
Abstract:Although there have been several recent advances in the application of deep learning algorithms to chest x-ray interpretation, we identify three major challenges for the translation of chest x-ray algorithms to the clinical setting. We examine the performance of the top 10 performing models on the CheXpert challenge leaderboard on three tasks: (1) TB detection, (2) pathology detection on photos of chest x-rays, and (3) pathology detection on data from an external institution. First, we find that the top 10 chest x-ray models on the CheXpert competition achieve an average AUC of 0.851 on the task of detecting TB on two public TB datasets without fine-tuning or including the TB labels in training data. Second, we find that the average performance of the models on photos of x-rays (AUC = 0.916) is similar to their performance on the original chest x-ray images (AUC = 0.924). Third, we find that the models tested on an external dataset either perform comparably to or exceed the average performance of radiologists. We believe that our investigation will inform rapid translation of deep learning algorithms to safe and effective clinical decision support tools that can be validated prospectively with large impact studies and clinical trials.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge