Andrew Johnston
Automated Structured Radiology Report Generation
May 30, 2025Abstract:Automated radiology report generation from chest X-ray (CXR) images has the potential to improve clinical efficiency and reduce radiologists' workload. However, most datasets, including the publicly available MIMIC-CXR and CheXpert Plus, consist entirely of free-form reports, which are inherently variable and unstructured. This variability poses challenges for both generation and evaluation: existing models struggle to produce consistent, clinically meaningful reports, and standard evaluation metrics fail to capture the nuances of radiological interpretation. To address this, we introduce Structured Radiology Report Generation (SRRG), a new task that reformulates free-text radiology reports into a standardized format, ensuring clarity, consistency, and structured clinical reporting. We create a novel dataset by restructuring reports using large language models (LLMs) following strict structured reporting desiderata. Additionally, we introduce SRR-BERT, a fine-grained disease classification model trained on 55 labels, enabling more precise and clinically informed evaluation of structured reports. To assess report quality, we propose F1-SRR-BERT, a metric that leverages SRR-BERT's hierarchical disease taxonomy to bridge the gap between free-text variability and structured clinical reporting. We validate our dataset through a reader study conducted by five board-certified radiologists and extensive benchmarking experiments.
Automated Real-time Assessment of Intracranial Hemorrhage Detection AI Using an Ensembled Monitoring Model (EMM)
May 16, 2025Abstract:Artificial intelligence (AI) tools for radiology are commonly unmonitored once deployed. The lack of real-time case-by-case assessments of AI prediction confidence requires users to independently distinguish between trustworthy and unreliable AI predictions, which increases cognitive burden, reduces productivity, and potentially leads to misdiagnoses. To address these challenges, we introduce Ensembled Monitoring Model (EMM), a framework inspired by clinical consensus practices using multiple expert reviews. Designed specifically for black-box commercial AI products, EMM operates independently without requiring access to internal AI components or intermediate outputs, while still providing robust confidence measurements. Using intracranial hemorrhage detection as our test case on a large, diverse dataset of 2919 studies, we demonstrate that EMM successfully categorizes confidence in the AI-generated prediction, suggesting different actions and helping improve the overall performance of AI tools to ultimately reduce cognitive burden. Importantly, we provide key technical considerations and best practices for successfully translating EMM into clinical settings.
Evaluating and Improving the Effectiveness of Synthetic Chest X-Rays for Medical Image Analysis
Nov 27, 2024Abstract:Purpose: To explore best-practice approaches for generating synthetic chest X-ray images and augmenting medical imaging datasets to optimize the performance of deep learning models in downstream tasks like classification and segmentation. Materials and Methods: We utilized a latent diffusion model to condition the generation of synthetic chest X-rays on text prompts and/or segmentation masks. We explored methods like using a proxy model and using radiologist feedback to improve the quality of synthetic data. These synthetic images were then generated from relevant disease information or geometrically transformed segmentation masks and added to ground truth training set images from the CheXpert, CANDID-PTX, SIIM, and RSNA Pneumonia datasets to measure improvements in classification and segmentation model performance on the test sets. F1 and Dice scores were used to evaluate classification and segmentation respectively. One-tailed t-tests with Bonferroni correction assessed the statistical significance of performance improvements with synthetic data. Results: Across all experiments, the synthetic data we generated resulted in a maximum mean classification F1 score improvement of 0.150453 (CI: 0.099108-0.201798; P=0.0031) compared to using only real data. For segmentation, the maximum Dice score improvement was 0.14575 (CI: 0.108267-0.183233; P=0.0064). Conclusion: Best practices for generating synthetic chest X-ray images for downstream tasks include conditioning on single-disease labels or geometrically transformed segmentation masks, as well as potentially using proxy modeling for fine-tuning such generations.
Overview of the First Shared Task on Clinical Text Generation: RRG24 and "Discharge Me!"
Sep 25, 2024



Abstract:Recent developments in natural language generation have tremendous implications for healthcare. For instance, state-of-the-art systems could automate the generation of sections in clinical reports to alleviate physician workload and streamline hospital documentation. To explore these applications, we present a shared task consisting of two subtasks: (1) Radiology Report Generation (RRG24) and (2) Discharge Summary Generation ("Discharge Me!"). RRG24 involves generating the 'Findings' and 'Impression' sections of radiology reports given chest X-rays. "Discharge Me!" involves generating the 'Brief Hospital Course' and 'Discharge Instructions' sections of discharge summaries for patients admitted through the emergency department. "Discharge Me!" submissions were subsequently reviewed by a team of clinicians. Both tasks emphasize the goal of reducing clinician burnout and repetitive workloads by generating documentation. We received 201 submissions from across 8 teams for RRG24, and 211 submissions from across 16 teams for "Discharge Me!".
* ACL Proceedings. BioNLP workshop
Detecting Underdiagnosed Medical Conditions with Deep Learning-Based Opportunistic CT Imaging
Sep 18, 2024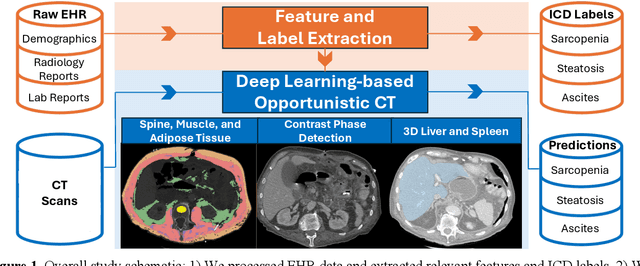
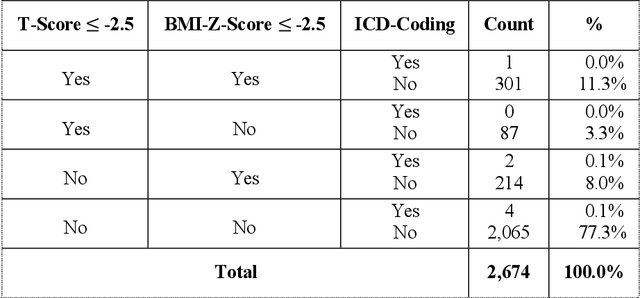
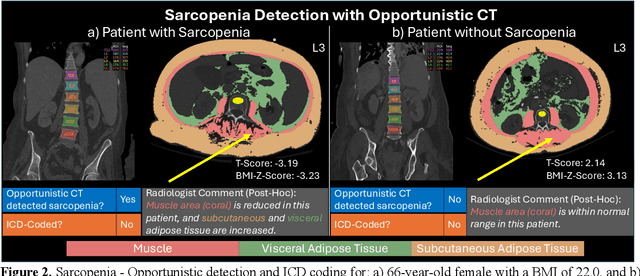
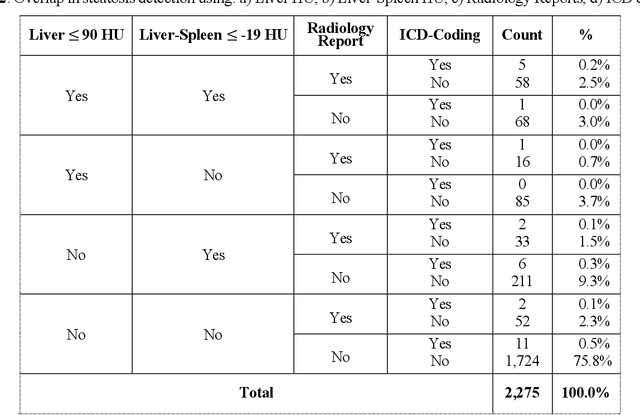
Abstract:Abdominal computed tomography (CT) scans are frequently performed in clinical settings. Opportunistic CT involves repurposing routine CT images to extract diagnostic information and is an emerging tool for detecting underdiagnosed conditions such as sarcopenia, hepatic steatosis, and ascites. This study utilizes deep learning methods to promote accurate diagnosis and clinical documentation. We analyze 2,674 inpatient CT scans to identify discrepancies between imaging phenotypes (characteristics derived from opportunistic CT scans) and their corresponding documentation in radiology reports and ICD coding. Through our analysis, we find that only 0.5%, 3.2%, and 30.7% of scans diagnosed with sarcopenia, hepatic steatosis, and ascites (respectively) through either opportunistic imaging or radiology reports were ICD-coded. Our findings demonstrate opportunistic CT's potential to enhance diagnostic precision and accuracy of risk adjustment models, offering advancements in precision medicine.
Merlin: A Vision Language Foundation Model for 3D Computed Tomography
Jun 10, 2024



Abstract:Over 85 million computed tomography (CT) scans are performed annually in the US, of which approximately one quarter focus on the abdomen. Given the current radiologist shortage, there is a large impetus to use artificial intelligence to alleviate the burden of interpreting these complex imaging studies. Prior state-of-the-art approaches for automated medical image interpretation leverage vision language models (VLMs). However, current medical VLMs are generally limited to 2D images and short reports, and do not leverage electronic health record (EHR) data for supervision. We introduce Merlin - a 3D VLM that we train using paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). We evaluate Merlin on 6 task types and 752 individual tasks. The non-adapted (off-the-shelf) tasks include zero-shot findings classification (31 findings), phenotype classification (692 phenotypes), and zero-shot cross-modal retrieval (image to findings and image to impressions), while model adapted tasks include 5-year disease prediction (6 diseases), radiology report generation, and 3D semantic segmentation (20 organs). We perform internal validation on a test set of 5,137 CTs, and external validation on 7,000 clinical CTs and on two public CT datasets (VerSe, TotalSegmentator). Beyond these clinically-relevant evaluations, we assess the efficacy of various network architectures and training strategies to depict that Merlin has favorable performance to existing task-specific baselines. We derive data scaling laws to empirically assess training data needs for requisite downstream task performance. Furthermore, unlike conventional VLMs that require hundreds of GPUs for training, we perform all training on a single GPU.
CheXagent: Towards a Foundation Model for Chest X-Ray Interpretation
Jan 22, 2024



Abstract:Chest X-rays (CXRs) are the most frequently performed imaging test in clinical practice. Recent advances in the development of vision-language foundation models (FMs) give rise to the possibility of performing automated CXR interpretation, which can assist physicians with clinical decision-making and improve patient outcomes. However, developing FMs that can accurately interpret CXRs is challenging due to the (1) limited availability of large-scale vision-language datasets in the medical image domain, (2) lack of vision and language encoders that can capture the complexities of medical data, and (3) absence of evaluation frameworks for benchmarking the abilities of FMs on CXR interpretation. In this work, we address these challenges by first introducing \emph{CheXinstruct} - a large-scale instruction-tuning dataset curated from 28 publicly-available datasets. We then present \emph{CheXagent} - an instruction-tuned FM capable of analyzing and summarizing CXRs. To build CheXagent, we design a clinical large language model (LLM) for parsing radiology reports, a vision encoder for representing CXR images, and a network to bridge the vision and language modalities. Finally, we introduce \emph{CheXbench} - a novel benchmark designed to systematically evaluate FMs across 8 clinically-relevant CXR interpretation tasks. Extensive quantitative evaluations and qualitative reviews with five expert radiologists demonstrate that CheXagent outperforms previously-developed general- and medical-domain FMs on CheXbench tasks. Furthermore, in an effort to improve model transparency, we perform a fairness evaluation across factors of sex, race and age to highlight potential performance disparities. Our project is at \url{https://stanford-aimi.github.io/chexagent.html}.
A Boosting Approach to Constructing an Ensemble Stack
Nov 28, 2022Abstract:An approach to evolutionary ensemble learning for classification is proposed in which boosting is used to construct a stack of programs. Each application of boosting identifies a single champion and a residual dataset, i.e. the training records that thus far were not correctly classified. The next program is only trained against the residual, with the process iterating until some maximum ensemble size or no further residual remains. Training against a residual dataset actively reduces the cost of training. Deploying the ensemble as a stack also means that only one classifier might be necessary to make a prediction, so improving interpretability. Benchmarking studies are conducted to illustrate competitiveness with the prediction accuracy of current state-of-the-art evolutionary ensemble learning algorithms, while providing solutions that are orders of magnitude simpler. Further benchmarking with a high cardinality dataset indicates that the proposed method is also more accurate and efficient than XGBoost.
Machine Learning for a Sustainable Energy Future
Oct 19, 2022Abstract:Transitioning from fossil fuels to renewable energy sources is a critical global challenge; it demands advances at the levels of materials, devices, and systems for the efficient harvesting, storage, conversion, and management of renewable energy. Researchers globally have begun incorporating machine learning (ML) techniques with the aim of accelerating these advances. ML technologies leverage statistical trends in data to build models for prediction of material properties, generation of candidate structures, optimization of processes, among other uses; as a result, they can be incorporated into discovery and development pipelines to accelerate progress. Here we review recent advances in ML-driven energy research, outline current and future challenges, and describe what is required moving forward to best lever ML techniques. To start, we give an overview of key ML concepts. We then introduce a set of key performance indicators to help compare the benefits of different ML-accelerated workflows for energy research. We discuss and evaluate the latest advances in applying ML to the development of energy harvesting (photovoltaics), storage (batteries), conversion (electrocatalysis), and management (smart grids). Finally, we offer an outlook of potential research areas in the energy field that stand to further benefit from the application of ML.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge