Michael Levin
Remapping and navigation of an embedding space via error minimization: a fundamental organizational principle of cognition in natural and artificial systems
Jan 20, 2026Abstract:The emerging field of diverse intelligence seeks an integrated view of problem-solving in agents of very different provenance, composition, and substrates. From subcellular chemical networks to swarms of organisms, and across evolved, engineered, and chimeric systems, it is hypothesized that scale-invariant principles of decision-making can be discovered. We propose that cognition in both natural and synthetic systems can be characterized and understood by the interplay between two equally important invariants: (1) the remapping of embedding spaces, and (2) the navigation within these spaces. Biological collectives, from single cells to entire organisms (and beyond), remap transcriptional, morphological, physiological, or 3D spaces to maintain homeostasis and regenerate structure, while navigating these spaces through distributed error correction. Modern Artificial Intelligence (AI) systems, including transformers, diffusion models, and neural cellular automata enact analogous processes by remapping data into latent embeddings and refining them iteratively through contextualization. We argue that this dual principle - remapping and navigation of embedding spaces via iterative error minimization - constitutes a substrate-independent invariant of cognition. Recognizing this shared mechanism not only illuminates deep parallels between living systems and artificial models, but also provides a unifying framework for engineering adaptive intelligence across scales.
Cognition spaces: natural, artificial, and hybrid
Jan 19, 2026Abstract:Cognitive processes are realized across an extraordinary range of natural, artificial, and hybrid systems, yet there is no unified framework for comparing their forms, limits, and unrealized possibilities. Here, we propose a cognition space approach that replaces narrow, substrate-dependent definitions with a comparative representation based on organizational and informational dimensions. Within this framework, cognition is treated as a graded capacity to sense, process, and act upon information, allowing systems as diverse as cells, brains, artificial agents, and human-AI collectives to be analyzed within a common conceptual landscape. We introduce and examine three cognition spaces -- basal aneural, neural, and human-AI hybrid -- and show that their occupation is highly uneven, with clusters of realized systems separated by large unoccupied regions. We argue that these voids are not accidental but reflect evolutionary contingencies, physical constraints, and design limitations. By focusing on the structure of cognition spaces rather than on categorical definitions, this approach clarifies the diversity of existing cognitive systems and highlights hybrid cognition as a promising frontier for exploring novel forms of complexity beyond those produced by biological evolution.
Advancing the Scientific Method with Large Language Models: From Hypothesis to Discovery
May 22, 2025Abstract:With recent Nobel Prizes recognising AI contributions to science, Large Language Models (LLMs) are transforming scientific research by enhancing productivity and reshaping the scientific method. LLMs are now involved in experimental design, data analysis, and workflows, particularly in chemistry and biology. However, challenges such as hallucinations and reliability persist. In this contribution, we review how Large Language Models (LLMs) are redefining the scientific method and explore their potential applications across different stages of the scientific cycle, from hypothesis testing to discovery. We conclude that, for LLMs to serve as relevant and effective creative engines and productivity enhancers, their deep integration into all steps of the scientific process should be pursued in collaboration and alignment with human scientific goals, with clear evaluation metrics. The transition to AI-driven science raises ethical questions about creativity, oversight, and responsibility. With careful guidance, LLMs could evolve into creative engines, driving transformative breakthroughs across scientific disciplines responsibly and effectively. However, the scientific community must also decide how much it leaves to LLMs to drive science, even when associations with 'reasoning', mostly currently undeserved, are made in exchange for the potential to explore hypothesis and solution regions that might otherwise remain unexplored by human exploration alone.
* 45 pages
What Lives? A meta-analysis of diverse opinions on the definition of life
May 19, 2025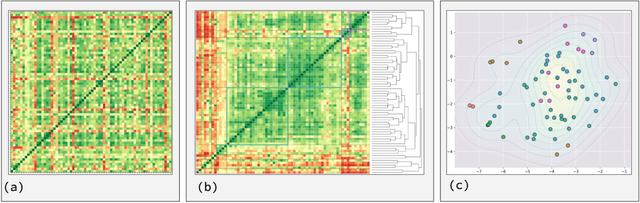
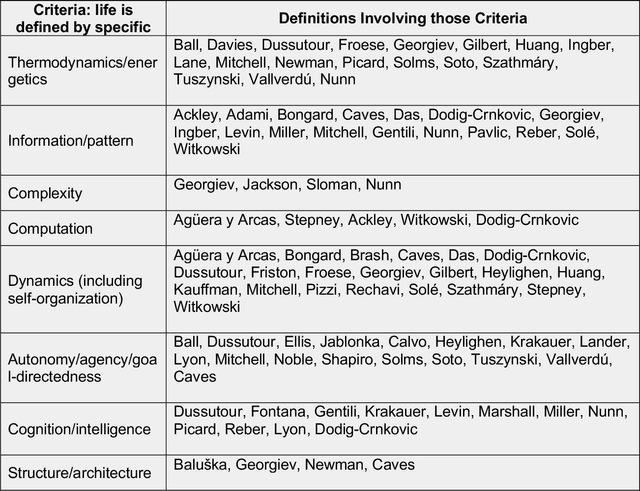
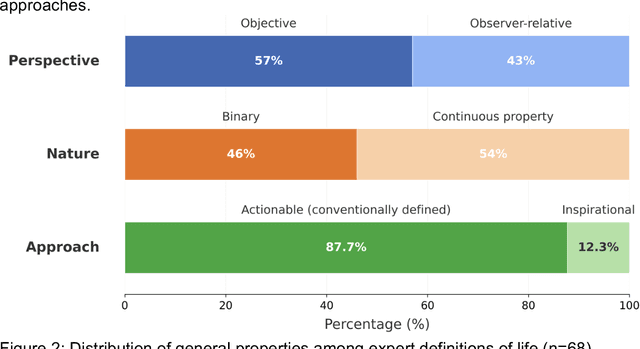
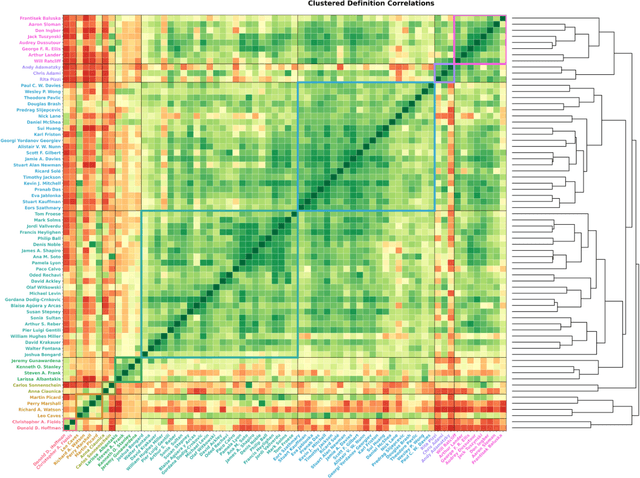
Abstract:The question of "what is life?" has challenged scientists and philosophers for centuries, producing an array of definitions that reflect both the mystery of its emergence and the diversity of disciplinary perspectives brought to bear on the question. Despite significant progress in our understanding of biological systems, psychology, computation, and information theory, no single definition for life has yet achieved universal acceptance. This challenge becomes increasingly urgent as advances in synthetic biology, artificial intelligence, and astrobiology challenge our traditional conceptions of what it means to be alive. We undertook a methodological approach that leverages large language models (LLMs) to analyze a set of definitions of life provided by a curated set of cross-disciplinary experts. We used a novel pairwise correlation analysis to map the definitions into distinct feature vectors, followed by agglomerative clustering, intra-cluster semantic analysis, and t-SNE projection to reveal underlying conceptual archetypes. This methodology revealed a continuous landscape of the themes relating to the definition of life, suggesting that what has historically been approached as a binary taxonomic problem should be instead conceived as differentiated perspectives within a unified conceptual latent space. We offer a new methodological bridge between reductionist and holistic approaches to fundamental questions in science and philosophy, demonstrating how computational semantic analysis can reveal conceptual patterns across disciplinary boundaries, and opening similar pathways for addressing other contested definitional territories across the sciences.
Giving Simulated Cells a Voice: Evolving Prompt-to-Intervention Models for Cellular Control
May 05, 2025



Abstract:Guiding biological systems toward desired states, such as morphogenetic outcomes, remains a fundamental challenge with far-reaching implications for medicine and synthetic biology. While large language models (LLMs) have enabled natural language as an interface for interpretable control in AI systems, their use as mediators for steering biological or cellular dynamics remains largely unexplored. In this work, we present a functional pipeline that translates natural language prompts into spatial vector fields capable of directing simulated cellular collectives. Our approach combines a large language model with an evolvable neural controller (Prompt-to-Intervention, or P2I), optimized via evolutionary strategies to generate behaviors such as clustering or scattering in a simulated 2D environment. We demonstrate that even with constrained vocabulary and simplified cell models, evolved P2I networks can successfully align cellular dynamics with user-defined goals expressed in plain language. This work offers a complete loop from language input to simulated bioelectric-like intervention to behavioral output, providing a foundation for future systems capable of natural language-driven cellular control.
Cancermorphic Computing Toward Multilevel Machine Intelligence
Mar 17, 2025Abstract:Despite their potential to address crucial bottlenecks in computing architectures and contribute to the pool of biological inspiration for engineering, pathological biological mechanisms remain absent from computational theory. We hereby introduce the concept of cancer-inspired computing as a paradigm drawing from the adaptive, resilient, and evolutionary strategies of cancer, for designing computational systems capable of thriving in dynamic, adversarial or resource-constrained environments. Unlike known bioinspired approaches (e.g., evolutionary and neuromorphic architectures), cancer-inspired computing looks at emulating the uniqueness of cancer cells survival tactics, such as somatic mutation, metastasis, angiogenesis and immune evasion, as parallels to desirable features in computing architectures, for example decentralized propagation and resource optimization, to impact areas like fault tolerance and cybersecurity. While the chaotic growth of cancer is currently viewed as uncontrollable in biology, randomness-based algorithms are already being successfully demonstrated in enhancing the capabilities of other computing architectures, for example chaos computing integration. This vision focuses on the concepts of multilevel intelligence and context-driven mutation, and their potential to simultaneously overcome plasticity-limited neuromorphic approaches and the randomness of chaotic approaches. The introduction of this concept aims to generate interdisciplinary discussion to explore the potential of cancer-inspired mechanisms toward powerful and resilient artificial systems.
Bio-inspired AI: Integrating Biological Complexity into Artificial Intelligence
Nov 22, 2024Abstract:The pursuit of creating artificial intelligence (AI) mirrors our longstanding fascination with understanding our own intelligence. From the myths of Talos to Aristotelian logic and Heron's inventions, we have sought to replicate the marvels of the mind. While recent advances in AI hold promise, singular approaches often fall short in capturing the essence of intelligence. This paper explores how fundamental principles from biological computation--particularly context-dependent, hierarchical information processing, trial-and-error heuristics, and multi-scale organization--can guide the design of truly intelligent systems. By examining the nuanced mechanisms of biological intelligence, such as top-down causality and adaptive interaction with the environment, we aim to illuminate potential limitations in artificial constructs. Our goal is to provide a framework inspired by biological systems for designing more adaptable and robust artificial intelligent systems.
Heuristically Adaptive Diffusion-Model Evolutionary Strategy
Nov 20, 2024



Abstract:Diffusion Models represent a significant advancement in generative modeling, employing a dual-phase process that first degrades domain-specific information via Gaussian noise and restores it through a trainable model. This framework enables pure noise-to-data generation and modular reconstruction of, images or videos. Concurrently, evolutionary algorithms employ optimization methods inspired by biological principles to refine sets of numerical parameters encoding potential solutions to rugged objective functions. Our research reveals a fundamental connection between diffusion models and evolutionary algorithms through their shared underlying generative mechanisms: both methods generate high-quality samples via iterative refinement on random initial distributions. By employing deep learning-based diffusion models as generative models across diverse evolutionary tasks and iteratively refining diffusion models with heuristically acquired databases, we can iteratively sample potentially better-adapted offspring parameters, integrating them into successive generations of the diffusion model. This approach achieves efficient convergence toward high-fitness parameters while maintaining explorative diversity. Diffusion models introduce enhanced memory capabilities into evolutionary algorithms, retaining historical information across generations and leveraging subtle data correlations to generate refined samples. We elevate evolutionary algorithms from procedures with shallow heuristics to frameworks with deep memory. By deploying classifier-free guidance for conditional sampling at the parameter level, we achieve precise control over evolutionary search dynamics to further specific genotypical, phenotypical, or population-wide traits. Our framework marks a major heuristic and algorithmic transition, offering increased flexibility, precision, and control in evolutionary optimization processes.
Diffusion Models are Evolutionary Algorithms
Oct 03, 2024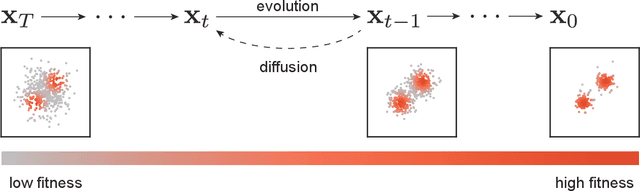
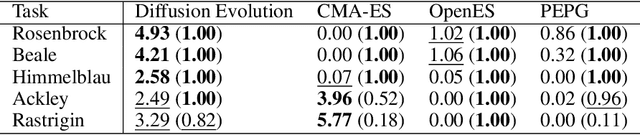
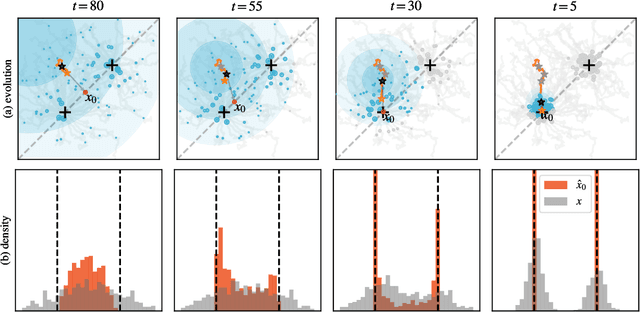
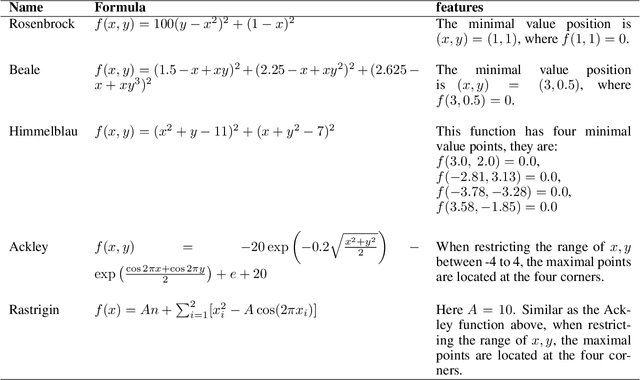
Abstract:In a convergence of machine learning and biology, we reveal that diffusion models are evolutionary algorithms. By considering evolution as a denoising process and reversed evolution as diffusion, we mathematically demonstrate that diffusion models inherently perform evolutionary algorithms, naturally encompassing selection, mutation, and reproductive isolation. Building on this equivalence, we propose the Diffusion Evolution method: an evolutionary algorithm utilizing iterative denoising -- as originally introduced in the context of diffusion models -- to heuristically refine solutions in parameter spaces. Unlike traditional approaches, Diffusion Evolution efficiently identifies multiple optimal solutions and outperforms prominent mainstream evolutionary algorithms. Furthermore, leveraging advanced concepts from diffusion models, namely latent space diffusion and accelerated sampling, we introduce Latent Space Diffusion Evolution, which finds solutions for evolutionary tasks in high-dimensional complex parameter space while significantly reducing computational steps. This parallel between diffusion and evolution not only bridges two different fields but also opens new avenues for mutual enhancement, raising questions about open-ended evolution and potentially utilizing non-Gaussian or discrete diffusion models in the context of Diffusion Evolution.
SBMLtoODEjax: efficient simulation and optimization of ODE SBML models in JAX
Jul 17, 2023


Abstract:Developing methods to explore, predict and control the dynamic behavior of biological systems, from protein pathways to complex cellular processes, is an essential frontier of research for bioengineering and biomedicine. Thus, significant effort has gone in computational inference and mathematical modeling of biological systems. This effort has resulted in the development of large collections of publicly-available models, typically stored and exchanged on online platforms (such as the BioModels Database) using the Systems Biology Markup Language (SBML), a standard format for representing mathematical models of biological systems. SBMLtoODEjax is a lightweight library that allows to automatically parse and convert SBML models into python models written end-to-end in JAX, a high-performance numerical computing library with automatic differentiation capabilities. SBMLtoODEjax is targeted at researchers that aim to incorporate SBML-specified ordinary differential equation (ODE) models into their python projects and machine learning pipelines, in order to perform efficient numerical simulation and optimization with only a few lines of code. SBMLtoODEjax is available at https://github.com/flowersteam/sbmltoodejax.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge